Transcripts expressed using a bicistronic vector pIREShyg2 are sensitized to nonsense-mediated mRNA decay
- PMID: 20513249
- PMCID: PMC2896932
- DOI: 10.1186/1471-2199-11-42
Transcripts expressed using a bicistronic vector pIREShyg2 are sensitized to nonsense-mediated mRNA decay
Abstract
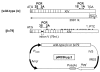
Background: pIREShyg2 has been widely used as a bicistronic expression vector. However, it is not known if the vector would affect the expression of cloned genes via nonsense-mediated mRNA decay (NMD), an mRNA surveillance system that degrades mRNA with a premature termination codon (PTC). In mammalian cells, the induction of NMD requires either a long 3'UTR or the presence of an exon-junction complex downstream of a PTC. The efficiency of NMD is greater when a PTC generates longer 3'UTR. pIREShyg2 provides the first cistron gene with a long 3'UTR consisting of a downstream intervening sequence (IVS), an internal ribosomal entry site (IRES) and the second cistron. Therefore, we hypothesized that the first cistron genes in pIREShyg2 are sensitized to NMD, which affects their expression levels. To examine this hypothesis, cDNAs encoding human granulocyte-macrophage colony-stimulating factor receptor beta chain (betac) and its splice variant (betac79), in which the retention of a 79-base intron caused a frameshift generating 18 PTCs, were cloned into pIREShyg2 and stably expressed in a murine cell line, Ba/F3.
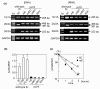
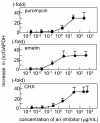
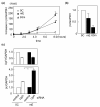
Results: Compared with wild-type betac, the mRNA levels of betac79 were less than one tenth and decayed faster. Both translation inhibition and Upf1 knockdown led to significantly greater up-regulation of betac79 than wild-type betac. However, the use of a monocistronic pMT21 vector abolished the up-regulatory effects of translation inhibition and Upf1 knockdown on both wild-type betac and betac79, suggesting that the NMD is attributable to a structural determinant in pIREShyg2. The elimination of the intron and the proximal 3' 17 PTCs did not alter the greater effects of translation inhibition on betac79, suggesting that the first PTC, which determines 3'UTR length, was sufficient to enhance NMD efficiency. Thus, transcripts of PTC-harboring genes with longer 3'UTR are more efficiently degraded by the vector-dependent NMD than those of wild-type genes with relatively shorter 3'UTR, resulting in minimized expression of truncated mutants.
Conclusions: We conclude that pIREShyg2, which sensitizes its bicistronic transcripts to NMD, may be useful for studying NMD but should be avoided when maximum expressions of PTC-harboring genes are required.
Figures

Similar articles
-
Chemical inhibition of eIF4A3 abolishes UPF1 recruitment onto mRNA encoding NMD factors and restores their expression.Biochem Biophys Res Commun. 2025 Feb 2;747:151270. doi: 10.1016/j.bbrc.2024.151270. Epub 2024 Dec 30. Biochem Biophys Res Commun. 2025. PMID: 39787791
-
The role of nucleotide composition in premature termination codon recognition.BMC Bioinformatics. 2016 Dec 7;17(1):519. doi: 10.1186/s12859-016-1384-z. BMC Bioinformatics. 2016. PMID: 27927164 Free PMC article.
-
A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay.PLoS Biol. 2008 Apr 29;6(4):e111. doi: 10.1371/journal.pbio.0060111. PLoS Biol. 2008. PMID: 18447585 Free PMC article.
-
Defining nonsense-mediated mRNA decay intermediates in human cells.Methods. 2019 Feb 15;155:68-76. doi: 10.1016/j.ymeth.2018.12.005. Epub 2018 Dec 19. Methods. 2019. PMID: 30576707 Free PMC article. Review.
-
Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells.Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):612-23. doi: 10.1016/j.bbagrm.2013.02.005. Epub 2013 Feb 20. Biochim Biophys Acta. 2013. PMID: 23435113 Review.
Cited by
-
Guidelines for minimal reporting requirements, design and interpretation of experiments involving the use of eukaryotic dual gene expression reporters (MINDR).Nat Struct Mol Biol. 2025 Mar;32(3):418-430. doi: 10.1038/s41594-025-01492-x. Epub 2025 Mar 3. Nat Struct Mol Biol. 2025. PMID: 40033152 Review.
-
Characterization of Optically and Electrically Evoked Dopamine Release in Striatal Slices from Digenic Knock-in Mice with DAT-Driven Expression of Channelrhodopsin.ACS Chem Neurosci. 2017 Feb 15;8(2):310-319. doi: 10.1021/acschemneuro.6b00300. Epub 2017 Feb 8. ACS Chem Neurosci. 2017. PMID: 28177213 Free PMC article.
-
Lipopolysaccharide inhibits myogenic differentiation of C2C12 myoblasts through the Toll-like receptor 4-nuclear factor-κB signaling pathway and myoblast-derived tumor necrosis factor-α.PLoS One. 2017 Jul 24;12(7):e0182040. doi: 10.1371/journal.pone.0182040. eCollection 2017. PLoS One. 2017. PMID: 28742154 Free PMC article.
-
Cell-Specific Targeting of Genetically Encoded Tools for Neuroscience.Annu Rev Genet. 2016 Nov 23;50:571-594. doi: 10.1146/annurev-genet-120215-035011. Epub 2016 Oct 6. Annu Rev Genet. 2016. PMID: 27732792 Free PMC article. Review.
-
Impairment of FOS mRNA stabilization following translation arrest in granulocytes from myelodysplastic syndrome patients.PLoS One. 2013 Apr 12;8(4):e61107. doi: 10.1371/journal.pone.0061107. Print 2013. PLoS One. 2013. PMID: 23593403 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

