Looping probabilities in model interphase chromosomes
- PMID: 20513384
- PMCID: PMC2877331
- DOI: 10.1016/j.bpj.2010.01.054
Looping probabilities in model interphase chromosomes
Abstract
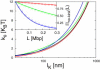
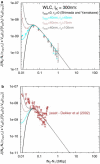
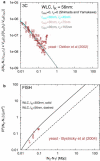
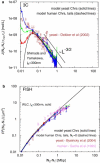
Fluorescence in-situ hybridization (FISH) and chromosome conformation capture (3C) are two powerful techniques for investigating the three-dimensional organization of the genome in interphase nuclei. The use of these techniques provides complementary information on average spatial distances (FISH) and contact probabilities (3C) for specific genomic sites. To infer the structure of the chromatin fiber or to distinguish functional interactions from random colocalization, it is useful to compare experimental data to predictions from statistical fiber models. The current estimates of the fiber stiffness derived from FISH and 3C differ by a factor of 5. They are based on the wormlike chain model and a heuristic modification of the Shimada-Yamakawa theory of looping for unkinkable, unconstrained, zero-diameter filaments. Here, we provide an extended theoretical and computational framework to explain the currently available experimental data for various species on the basis of a unique, minimal model of decondensing chromosomes: a kinkable, topologically constraint, semiflexible polymer with the (FISH) Kuhn length of l(K) = 300 nm, 10 kinks per Mbp, and a contact distance of 45 nm. In particular: 1), we reconsider looping of finite-diameter filaments on the basis of an analytical approximation (novel, to our knowledge) of the wormlike chain radial density and show that unphysically large contact radii would be required to explain the 3C data based on the FISH estimate of the fiber stiffness; 2), we demonstrate that the observed interaction frequencies at short genomic lengths can be explained by the presence of a low concentration of curvature defects (kinks); and 3), we show that the most recent experimental 3C data for human chromosomes are in quantitative agreement with interaction frequencies extracted from our simulations of topologically confined model chromosomes.
Copyright (c) 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



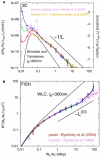
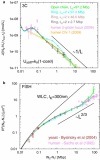
Similar articles
-
Simulation of different three-dimensional polymer models of interphase chromosomes compared to experiments-an evaluation and review framework of the 3D genome organization.Semin Cell Dev Biol. 2019 Jun;90:19-42. doi: 10.1016/j.semcdb.2018.07.012. Epub 2018 Aug 24. Semin Cell Dev Biol. 2019. PMID: 30125668 Review.
-
Simulation of Different Three-Dimensional Models of Whole Interphase Nuclei Compared to Experiments - A Consistent Scale-Bridging Simulation Framework for Genome Organization.Results Probl Cell Differ. 2022;70:495-549. doi: 10.1007/978-3-031-06573-6_18. Results Probl Cell Differ. 2022. PMID: 36348120
-
Lattice animal model of chromosome organization.Phys Rev E Stat Nonlin Soft Matter Phys. 2012 Jul;86(1 Pt 1):011911. doi: 10.1103/PhysRevE.86.011911. Epub 2012 Jul 12. Phys Rev E Stat Nonlin Soft Matter Phys. 2012. PMID: 23005456
-
Models that include supercoiling of topological domains reproduce several known features of interphase chromosomes.Nucleic Acids Res. 2014 Mar;42(5):2848-55. doi: 10.1093/nar/gkt1353. Epub 2013 Dec 23. Nucleic Acids Res. 2014. PMID: 24366878 Free PMC article.
-
Chromatin Domains: The Unit of Chromosome Organization.Mol Cell. 2016 Jun 2;62(5):668-80. doi: 10.1016/j.molcel.2016.05.018. Mol Cell. 2016. PMID: 27259200 Free PMC article. Review.
Cited by
-
Entropy gives rise to topologically associating domains.Nucleic Acids Res. 2016 Jul 8;44(12):5540-9. doi: 10.1093/nar/gkw510. Epub 2016 Jun 2. Nucleic Acids Res. 2016. PMID: 27257057 Free PMC article.
-
The post-mitotic state in neurons correlates with a stable nuclear higher-order structure.Commun Integr Biol. 2012 Mar 1;5(2):134-9. doi: 10.4161/cib.18761. Commun Integr Biol. 2012. PMID: 22808316 Free PMC article.
-
Mechanisms of Chromosome Folding and Nuclear Organization: Their Interplay and Open Questions.Cold Spring Harb Perspect Biol. 2022 Jul 1;14(7):a040147. doi: 10.1101/cshperspect.a040147. Cold Spring Harb Perspect Biol. 2022. PMID: 34518339 Free PMC article. Review.
-
Organization of the mitotic chromosome.Science. 2013 Nov 22;342(6161):948-53. doi: 10.1126/science.1236083. Epub 2013 Nov 7. Science. 2013. PMID: 24200812 Free PMC article.
-
Closing the loop: 3C versus DNA FISH.Genome Biol. 2016 Oct 19;17(1):215. doi: 10.1186/s13059-016-1081-2. Genome Biol. 2016. PMID: 27760553 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

