SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes
- PMID: 20513433
- PMCID: PMC3161732
- DOI: 10.1016/j.molcel.2010.02.040
SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes
Abstract
The ATP-dependent chromatin remodeling complex SWI/SNF regulates transcription and has been implicated in promoter nucleosome eviction. Efficient nucleosome disassembly by SWI/SNF alone in biochemical assays, however, has not been directly observed. Employing a model system of dinucleosomes rather than mononucleosomes, we demonstrate that remodeling leads to ordered and efficient disassembly of one of the two nucleosomes. An H2A/H2B dimer is first rapidly displaced, and then, in a slower reaction, an entire histone octamer is lost. Nucleosome disassembly by SWI/SNF did not require additional factors such as chaperones or acceptors of histones. Observations in single molecules as well as bulk measurement suggest that a key intermediate in this process is one in which a nucleosome is moved toward the adjacent nucleosome. SWI/SNF recruited by the transcriptional activator Gal4-VP16 preferentially mobilizes the proximal nucleosome and destabilizes the adjacent nucleosome.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
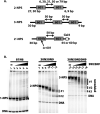


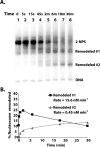
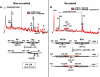


Comment in
-
When push comes to shove: SWI/SNF uses a nucleosome to get rid of a nucleosome.Mol Cell. 2010 May 28;38(4):484-6. doi: 10.1016/j.molcel.2010.05.005. Mol Cell. 2010. PMID: 20513424 Free PMC article.
References
-
- Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell. 2004;14:657–666. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

