Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation
- PMID: 20513434
- PMCID: PMC5975946
- DOI: 10.1016/j.molcel.2010.03.016
Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation
Abstract
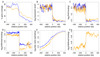
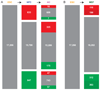
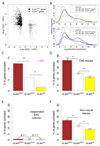
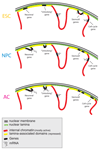
The three-dimensional organization of chromosomes within the nucleus and its dynamics during differentiation are largely unknown. To visualize this process in molecular detail, we generated high-resolution maps of genome-nuclear lamina interactions during subsequent differentiation of mouse embryonic stem cells via lineage-committed neural precursor cells into terminally differentiated astrocytes. This reveals that a basal chromosome architecture present in embryonic stem cells is cumulatively altered at hundreds of sites during lineage commitment and subsequent terminal differentiation. This remodeling involves both individual transcription units and multigene regions and affects many genes that determine cellular identity. Often, genes that move away from the lamina are concomitantly activated; many others, however, remain inactive yet become unlocked for activation in a next differentiation step. These results suggest that lamina-genome interactions are widely involved in the control of gene expression programs during lineage commitment and terminal differentiation.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures



Comment in
-
The silence of the LADs: dynamic genome-lamina interactions during ESC differentiation.Cell Stem Cell. 2010 Jun 4;6(6):495-7. doi: 10.1016/j.stem.2010.05.006. Cell Stem Cell. 2010. PMID: 20569682 No abstract available.
References
-
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

