Insulin-like growth factor-binding protein-3 promotes transforming growth factor-{beta}1-mediated epithelial-to-mesenchymal transition and motility in transformed human esophageal cells
- PMID: 20513670
- PMCID: PMC2915630
- DOI: 10.1093/carcin/bgq108
Insulin-like growth factor-binding protein-3 promotes transforming growth factor-{beta}1-mediated epithelial-to-mesenchymal transition and motility in transformed human esophageal cells
Abstract
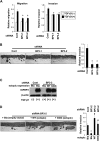
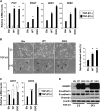
Insulin-like growth factor-binding protein (IGFBP)-3 is overexpressed frequently in esophageal squamous cell carcinoma. Yet, the role of IGFBP3 in esophageal tumor biology remains to be elucidated. We find that IGFBP3 facilitates transforming growth factor (TGF)-beta1-mediated epithelial-to-mesenchymal transition (EMT) in transformed human esophageal epithelial cells, EPC2-hTERT-EGFR-p53(R175H). In organotypic 3D culture, a form of human tissue engineering, laser-capture microdissection revealed concurrent upregulation of TGF-beta target genes, IGFBP3 and EMT-related genes in the cells invading into the stromal compartment. IGFBP3 enhanced TGF-beta1-mediated EMT as well as transcription factors essential in EMT by allowing persistent SMAD2 and SMAD3 phosphorylation. TGF-beta1-mediated EMT and cell invasion were enhanced by ectopically expressed IGFBP3 and suppressed by RNA interference directed against IGFBP3. The IGFBP3 knockdown effect was rescued by IGFBP3(I56G/L80G/L81G), a mutant IGFBP3 lacking an insulin-like growth factor (IGF)-binding capacity. Thus, IGFBP3 can regulate TGF-beta1-mediated EMT and cell invasion in an IGF or insulin-like growth factor 1 receptor-independent manner. IGFBP3(I56G/L80G/L81G) also promoted EMT in vivo in a Ras-transformed human esophageal cell line T-TeRas upon xenograft transplantation in nude mice. In aggregate, IGFBP3 may have a novel IGF-binding independent biological function in regulation of TGF-beta1-mediated EMT and cell invasion.
Figures




References
-
- Enzinger PC, et al. Esophageal cancer. N. Engl. J. Med. 2003;349:2241–2252. - PubMed
-
- Nakagawa H, et al. Biology of esophageal cancer. In: Rustgi AK, editor. Gastrointestinal Cancers. London: Elsevier; 2003. pp. 241–251.
-
- Itakura Y, et al. Epidermal growth factor receptor overexpression in esophageal carcinoma. An immunohistochemical study correlated with clinicopathologic findings and DNA amplification. Cancer. 1994;74:795–804. - PubMed
-
- Parenti AR, et al. p53 overexpression in the multistep process of esophageal carcinogenesis. Am. J. Surg. Pathol. 1995;19:1418–1422. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

