Constant pH replica exchange molecular dynamics in biomolecules using a discrete protonation model
- PMID: 20514364
- PMCID: PMC2877402
- DOI: 10.1021/ct900676b
Constant pH replica exchange molecular dynamics in biomolecules using a discrete protonation model
Abstract
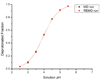
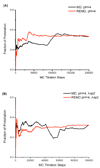
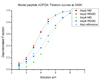
A constant pH replica exchange molecular dynamics (REMD) method is proposed and implemented to improve coupled protonation and conformational state sampling. By mixing conformational sampling at constant pH (with discrete protonation states) with a temperature ladder, this method avoids conformational trapping. Our method was tested and applied to seven different biological systems. The constant pH REMD not only predicted pKa correctly for small, model compounds but also converged faster than constant pH molecular dynamics (MD). We further tested our constant pH REMD on a heptapeptide from ovomucoid third domain (OMTKY3). Although constant pH REMD and MD produced very close pKa values, the constant pH REMD showed its advantage in the efficiency of conformational and protonation state samplings.
Figures


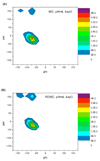
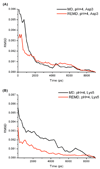
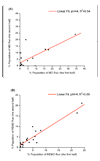
References
Grants and funding
LinkOut - more resources
Full Text Sources
