Nuclear expression of FLT1 and its ligand PGF in FUS-DDIT3 carrying myxoid liposarcomas suggests the existence of an intracrine signaling loop
- PMID: 20515481
- PMCID: PMC2889895
- DOI: 10.1186/1471-2407-10-249
Nuclear expression of FLT1 and its ligand PGF in FUS-DDIT3 carrying myxoid liposarcomas suggests the existence of an intracrine signaling loop
Abstract
Background: The FUS-DDIT3 fusion oncogene encodes an abnormal transcription factor that has a causative role in the development of myxoid/round-cell liposarcomas (MLS/RCLS). We have previously identified FLT1 (VEGFR1) as a candidate downstream target gene of FUS-DDIT3. The aim of this study was to investigate expression of FLT1 and its ligands in MLS cells.
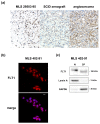
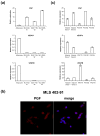
Methods: HT1080 human fibrosarcoma cells were transiently transfected with FUS-DDIT3-GFP variant constructs and FLT1 expression was measured by quantitative real-time PCR. In addition, FLT1, PGF, VEGFA and VEGFB expression was measured in MLS/RCLS cell lines, MLS/RCLS tumors and in normal adiopocytes. We analyzed nine cases of MLS/RCLS and one cell line xenografted in mice for FLT1 protein expression using immunohistochemistry. MLS/RCLS cell lines were also analyzed for FLT1 by immunofluorescence and western blot. MLS/RCLS cell lines were additionally treated with FLT1 tyrosine kinase inhibitors and assayed for alterations in proliferation rate.
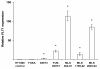
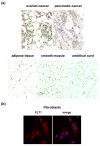
Results: FLT1 expression was dramatically increased in transfected cells stably expressing FUS-DDIT3 and present at high levels in cell lines derived from MLS. The FLT1 protein showed a strong nuclear expression in cells of MLS tissue as well as in cultured MLS cells, which was confirmed by cellular fractionation. Tissue array analysis showed a nuclear expression of the FLT1 protein also in several other tumor and normal cell types including normal adipocytes. The FLT1 ligand coding gene PGF was highly expressed in cultured MLS cells compared to normal adipocytes while the other ligand genes VEGFA and VEGFB were expressed to lower levels. A more heterogeneous expression pattern of these genes were observed in tumor samples. No changes in proliferation rate of MLS cells were detected at concentrations for which the kinase inhibitors have shown specific inhibition of FLT1.
Conclusions: Our results imply that FLT1 is induced as an indirect downstream effect of FUS-DDIT3 expression in MLS. This could be a consequence of the ability of FUS-DDIT3 to hijack parts of normal adipose tissue development and reprogram primary cells to a liposarcoma-like phenotype. The findings of nuclear FLT1 protein and expression of corresponding ligands in MLS and normal tissues may have implications for tissue homeostasis and tumor development through auto- or intracrine signaling.
Figures
References
-
- Fletcher CDM, Unni KK, Mertens F. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Soft Tissue and Bone. IARC Press: Lyon; 2002.
-
- Andersson MK, Stahlberg A, Arvidsson Y, Olofsson A, Semb H, Stenman G, Nilsson O, Aman P. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 2008;9:37. doi: 10.1186/1471-2121-9-37. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

