Novel polymorphisms of nuclear receptor SHP associated with functional and structural changes
- PMID: 20516075
- PMCID: PMC2915723
- DOI: 10.1074/jbc.M110.133280
Novel polymorphisms of nuclear receptor SHP associated with functional and structural changes
Abstract
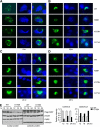
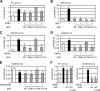
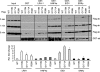
We identified three heterozygous nonsynonymous single nucleotide polymorphisms in the small heterodimer partner (SHP, NROB2) gene in normal subjects and CADASIL (cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy)-like patients, including two novel missense mutations (p.R38H, p.K170N) and one of the previously reported polymorphism (p.G171A). Four novel heterozygous mutations were also identified in the intron ((Intron)1265T-->A), 3'-untranslated region ((3'-UTR)101C-->G, (3'-UTR)186T-->C), and promoter ((Pro)-423C-->T) of the SHP gene. The exonic R38H and K170N mutants exhibited impaired nuclear translocation. K170N made SHP more susceptible to ubiquitination mediated degradation and blocked SHP acetylation, which displayed lost repressive activity on its interacting partners ERRgamma and HNF4alpha but not LRH-1. In contrast, G171A increased SHP mRNA and protein expression and maintained normal function. In general, the interaction of SHP mutants with LRH-1 and EID1 was enhanced. K170N also markedly impaired the recruitment of SHP, HNF4alpha, HDAC1, and HDAC3 to the apoCIII promoter. Molecular dynamics simulations of SHP showed that G171A stabilized the nuclear receptor boxes, whereas K170N promoted the conformational destabilization of all the structural elements of the receptor. This study suggests that genetic variations in SHP are common among human subjects and the Lys-170 residue plays a key role in controlling SHP ubiquitination and acetylation associated with SHP protein stability and repressive function.
Figures





Similar articles
-
Differential regulation of the orphan nuclear receptor small heterodimer partner (SHP) gene promoter by orphan nuclear receptor ERR isoforms.J Biol Chem. 2002 Jan 18;277(3):1739-48. doi: 10.1074/jbc.M106140200. Epub 2001 Nov 8. J Biol Chem. 2002. PMID: 11705994
-
Nuclear receptor SHP activates miR-206 expression via a cascade dual inhibitory mechanism.PLoS One. 2009 Sep 1;4(9):e6880. doi: 10.1371/journal.pone.0006880. PLoS One. 2009. PMID: 19721712 Free PMC article.
-
Contribution of variants in the small heterodimer partner gene to birthweight, adiposity, and insulin levels: mutational analysis and association studies in multiple populations.Diabetes. 2003 May;52(5):1288-91. doi: 10.2337/diabetes.52.5.1288. Diabetes. 2003. PMID: 12716767
-
A Novel Heterozygous Variant in Exon 19 of NOTCH3 in a Saudi Family with Cerebral Autosomal Dominant Arteriopathy with Subcortical Infarcts and Leukoencephalopathy.J Stroke Cerebrovasc Dis. 2020 Jul;29(7):104832. doi: 10.1016/j.jstrokecerebrovasdis.2020.104832. Epub 2020 May 13. J Stroke Cerebrovasc Dis. 2020. PMID: 32414585 Review.
-
Structure and function of the atypical orphan nuclear receptor small heterodimer partner.Int Rev Cytol. 2007;261:117-58. doi: 10.1016/S0074-7696(07)61003-1. Int Rev Cytol. 2007. PMID: 17560281 Review.
Cited by
-
Cross-regulation of protein stability by p53 and nuclear receptor SHP.PLoS One. 2012;7(6):e39789. doi: 10.1371/journal.pone.0039789. Epub 2012 Jun 21. PLoS One. 2012. PMID: 22737255 Free PMC article.
-
Genome-wide transcriptome analysis identifies novel gene signatures implicated in human chronic liver disease.Am J Physiol Gastrointest Liver Physiol. 2013 Sep 1;305(5):G364-74. doi: 10.1152/ajpgi.00077.2013. Epub 2013 Jun 27. Am J Physiol Gastrointest Liver Physiol. 2013. PMID: 23812039 Free PMC article.
-
H19 promotes cholestatic liver fibrosis by preventing ZEB1-mediated inhibition of epithelial cell adhesion molecule.Hepatology. 2017 Oct;66(4):1183-1196. doi: 10.1002/hep.29209. Epub 2017 Aug 26. Hepatology. 2017. PMID: 28407375 Free PMC article.
-
Repression of hepatocyte nuclear factor 4 alpha by AP-1 underlies dyslipidemia associated with retinoic acid.J Lipid Res. 2019 Apr;60(4):794-804. doi: 10.1194/jlr.M088880. Epub 2019 Feb 1. J Lipid Res. 2019. PMID: 30709899 Free PMC article.
-
Zinc-induced Dnmt1 expression involves antagonism between MTF-1 and nuclear receptor SHP.Nucleic Acids Res. 2012 Jun;40(11):4850-60. doi: 10.1093/nar/gks159. Epub 2012 Feb 22. Nucleic Acids Res. 2012. PMID: 22362755 Free PMC article.
References
-
- Markward N. J. (2007) AMIA Annual Symposium Proceedings, Chicago, IL, November 10–14, 2007, p. 1041, American Medical Informatics Association, Bethesda, MD - PubMed
-
- Lee H. K., Lee Y. K., Park S. H., Kim Y. S., Park S. H., Lee J. W., Kwon H. B., Soh J., Moore D. D., Choi H. S. (1998) J. Biol. Chem. 273, 14398–14402 - PubMed
-
- Wang L., Liu J., Saha P., Huang J., Chan L., Spiegelman B., Moore D. D. (2005) Cell Metab. 2, 227–238 - PubMed
-
- Huang J., Iqbal J., Saha P. K., Liu J., Chan L., Hussain M. M., Moore D. D., Wang L. (2007) Hepatology 46, 147–157 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

