Affinity purification of the Arabidopsis 26 S proteasome reveals a diverse array of plant proteolytic complexes
- PMID: 20516081
- PMCID: PMC2919120
- DOI: 10.1074/jbc.M110.136622
Affinity purification of the Arabidopsis 26 S proteasome reveals a diverse array of plant proteolytic complexes
Abstract
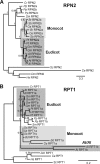
Selective proteolysis in plants is largely mediated by the ubiquitin (Ub)/proteasome system in which substrates, marked by the covalent attachment of Ub, are degraded by the 26 S proteasome. The 26 S proteasome is composed of two subparticles, the 20 S core protease (CP) that compartmentalizes the protease active sites and the 19 S regulatory particle that recognizes and translocates appropriate substrates into the CP lumen for breakdown. Here, we describe an affinity method to rapidly purify epitope-tagged 26 S proteasomes intact from Arabidopsis thaliana. In-depth mass spectrometric analyses of preparations generated from young seedlings confirmed that the 2.5-MDa CP-regulatory particle complex is actually a heterogeneous set of particles assembled with paralogous pairs for most subunits. A number of these subunits are modified post-translationally by proteolytic processing, acetylation, and/or ubiquitylation. Several proteasome-associated proteins were also identified that likely assist in complex assembly and regulation. In addition, we detected a particle consisting of the CP capped by the single subunit PA200 activator that may be involved in Ub-independent protein breakdown. Taken together, it appears that a diverse and highly dynamic population of proteasomes is assembled in plants, which may expand the target specificity and functions of intracellular proteolysis.
Figures

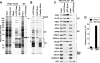




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

