Constructing partial models of cells
- PMID: 20516136
- PMCID: PMC2869526
- DOI: 10.1101/cshperspect.a004945
Constructing partial models of cells
Abstract
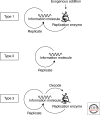
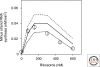
Understanding the origin of life requires knowledge not only of the origin of biological molecules such as amino acids, nucleotides and their polymers, but also the manner in which those molecules are integrated into the organized systems that characterize cellular life. In this article, we introduce a constructive approach to understand how biological molecules can be arranged to achieve a higher-order biological function: replication of genetic information.
Figures

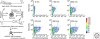

References
-
- Bartel DP, Szostak JW 1993. Isolation of new ribozymes from a large pool of random sequences. Science 261:1411–1418 - PubMed
-
- Biebricher CK, Eigen M, Gardiner WC Jr 1985. Kinetics of RNA replication: competition and selection among self-replicating RNA species. Biochemistry 24:6550–6560 - PubMed
-
- Chakrabarti AC, Breaker RR, Joyce GF, Deamer DW 1994. Production of RNA by a polymerase protein encapsulated within phospholipid vesicles. J Mol Evol 39:555–559 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
