HIV-1 Rev-binding protein accelerates cellular uptake of iron to drive Notch-induced T cell leukemogenesis in mice
- PMID: 20516639
- PMCID: PMC2898592
- DOI: 10.1172/JCI41277
HIV-1 Rev-binding protein accelerates cellular uptake of iron to drive Notch-induced T cell leukemogenesis in mice
Abstract
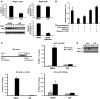
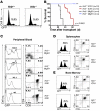
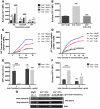
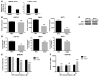
Somatic activating mutations in Notch1 contribute to the pathogenesis of T cell acute lymphoblastic lymphoma (T-ALL), but how activated Notch1 signaling exerts this oncogenic effect is not completely understood. Here we identify HIV-1 Rev-binding protein (Hrb), a component of the clathrin-mediated endocytosis machinery, as a critical mediator of Notch-induced T-ALL development in mice. Hrb was found to be a direct transcriptional target of Notch1, and Hrb loss reduced the incidence or delayed the onset of T-ALL in mouse models in which activated Notch1 signaling either contributes to or drives leukemogenesis. Consistent with this observation, Hrb supported survival and proliferation of hematopoietic and T cell precursor cells in vitro. We demonstrated that Hrb accelerated the uptake of transferrin, which was required for upregulation of the T cell protooncogene p21. Indeed, iron-deficient mice developed Notch1-induced T-ALL substantially more slowly than control mice, further supporting a critical role for iron uptake during leukemogenesis. Taken together, these results reveal that Hrb is a critical Notch target gene that mediates lymphoblast transformation and disease progression via its ability to satisfy the enhanced demands of transformed lymphoblasts for iron. Further, our data suggest that Hrb may be targeted to improve current treatment or design novel therapies for human T-ALL patients.
Figures

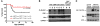
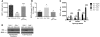
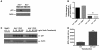

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

