Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo
- PMID: 20517297
- PMCID: PMC2905244
- DOI: 10.1038/emboj.2010.106
Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo
Abstract
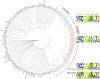
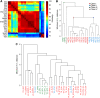
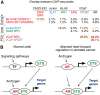
Members of the large ETS family of transcription factors (TFs) have highly similar DNA-binding domains (DBDs)-yet they have diverse functions and activities in physiology and oncogenesis. Some differences in DNA-binding preferences within this family have been described, but they have not been analysed systematically, and their contributions to targeting remain largely uncharacterized. We report here the DNA-binding profiles for all human and mouse ETS factors, which we generated using two different methods: a high-throughput microwell-based TF DNA-binding specificity assay, and protein-binding microarrays (PBMs). Both approaches reveal that the ETS-binding profiles cluster into four distinct classes, and that all ETS factors linked to cancer, ERG, ETV1, ETV4 and FLI1, fall into just one of these classes. We identify amino-acid residues that are critical for the differences in specificity between all the classes, and confirm the specificities in vivo using chromatin immunoprecipitation followed by sequencing (ChIP-seq) for a member of each class. The results indicate that even relatively small differences in in vitro binding specificity of a TF contribute to site selectivity in vivo.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Similar articles
-
Structures of the Ets Protein DNA-binding Domains of Transcription Factors Etv1, Etv4, Etv5, and Fev: DETERMINANTS OF DNA BINDING AND REDOX REGULATION BY DISULFIDE BOND FORMATION.J Biol Chem. 2015 May 29;290(22):13692-709. doi: 10.1074/jbc.M115.646737. Epub 2015 Apr 12. J Biol Chem. 2015. PMID: 25866208 Free PMC article.
-
Electrostatic repulsion causes anticooperative DNA binding between tumor suppressor ETS transcription factors and JUN-FOS at composite DNA sites.J Biol Chem. 2018 Nov 30;293(48):18624-18635. doi: 10.1074/jbc.RA118.003352. Epub 2018 Oct 12. J Biol Chem. 2018. PMID: 30315111 Free PMC article.
-
Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family.Genes Dev. 2007 Aug 1;21(15):1882-94. doi: 10.1101/gad.1561707. Epub 2007 Jul 24. Genes Dev. 2007. PMID: 17652178 Free PMC article.
-
Genomic and biochemical insights into the specificity of ETS transcription factors.Annu Rev Biochem. 2011;80:437-71. doi: 10.1146/annurev.biochem.79.081507.103945. Annu Rev Biochem. 2011. PMID: 21548782 Free PMC article. Review.
-
ETS transcription factors as emerging drug targets in cancer.Med Res Rev. 2020 Jan;40(1):413-430. doi: 10.1002/med.21575. Epub 2019 Mar 29. Med Res Rev. 2020. PMID: 30927317 Review.
Cited by
-
ETS variant 1 regulates matrix metalloproteinase-7 transcription in LNCaP prostate cancer cells.Oncol Rep. 2013 Jan;29(1):306-14. doi: 10.3892/or.2012.2079. Epub 2012 Oct 16. Oncol Rep. 2013. PMID: 23076342 Free PMC article.
-
Annotation of functional variation in personal genomes using RegulomeDB.Genome Res. 2012 Sep;22(9):1790-7. doi: 10.1101/gr.137323.112. Genome Res. 2012. PMID: 22955989 Free PMC article.
-
Inferring dynamic gene regulatory networks in cardiac differentiation through the integration of multi-dimensional data.BMC Bioinformatics. 2015 Mar 7;16:74. doi: 10.1186/s12859-015-0460-0. BMC Bioinformatics. 2015. PMID: 25887857 Free PMC article.
-
Intracellular and Intercellular Gene Regulatory Network Inference From Time-Course Individual RNA-Seq.Front Bioinform. 2021 Nov 11;1:777299. doi: 10.3389/fbinf.2021.777299. eCollection 2021. Front Bioinform. 2021. PMID: 36303726 Free PMC article.
-
The ETS transcription factors ELK1 and GABPA regulate different gene networks to control MCF10A breast epithelial cell migration.PLoS One. 2012;7(12):e49892. doi: 10.1371/journal.pone.0049892. Epub 2012 Dec 20. PLoS One. 2012. PMID: 23284628 Free PMC article.
References
-
- Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7: 986–995 - PubMed
-
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 - PubMed
-
- Bartel FO, Higuchi T, Spyropoulos DD (2000) Mouse models in the study of the Ets family of transcription factors. Oncogene 19: 6443–6454 - PubMed
-
- Batchelor AH, Piper DE, de la Brousse FC, McKnight SL, Wolberger C (1998) The structure of GABPalpha/beta: an ETS domain-ankyrin repeat heterodimer bound to DNA. Science 279: 1037–1041 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

