Neuropathogenic SIVsmmFGb genetic diversity and selection-induced tissue-specific compartmentalization during chronic infection and temporal evolution of viral genes in lymphoid tissues and regions of the central nervous system
- PMID: 20518690
- PMCID: PMC2932553
- DOI: 10.1089/aid.2009.0168
Neuropathogenic SIVsmmFGb genetic diversity and selection-induced tissue-specific compartmentalization during chronic infection and temporal evolution of viral genes in lymphoid tissues and regions of the central nervous system
Abstract
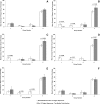
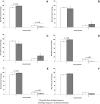
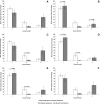
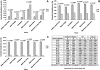
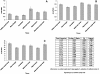
SIVsmmFGb is a lentivirus swarm that induces neuropathology in over 90% of infected pigtailed macaques and reliably models central nervous system HIV infection in people. We have previously studied SIVsmmFGb genetic diversity and compartmentalization during acute infection, but little is understood about diversity and intertissue compartmentalization during chronic infection. Tissue-specific pressure appeared to affect the diversity of Nef sequences between tissues, but changes to the Env V1 region and Int diversity were similar across all tissues. At 2 months postinfection, compartmentalization of the SIVsmmFGb env V1 region, nef, and int was noted between different brain regions and between brain regions and lymph nodes. Convergent evolution of the nef and env V1 region, and divergent evolution of int, was noted between compartments and all genes demonstrated intratissue temporal segregation. For the env V1 region and nef, temporal segregation was stronger in the brain regions than the periphery, but little difference between tissues was noted for int. Positive selection of the env V1 region appeared in most tissues at 2 months postinfection, whereas nef and int faced negative selection in all tissues. Positive selection of the env V1 region sequences increased in some brain regions over time. SIVsmmFGb nef and int sequences each saw increased negative selection in brain regions, and one lymph node, over the course of infection. Functional differences between tissue compartments decreased over time for int and env V1 region sequences, but increased for nef sequences.
Figures
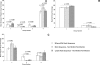


References
-
- UNAIDS. AIDS Epidemic Update. 2007.
-
- Glenn AA. Novembre FJ. A single amino acid change in gp41 is linked to the macrophage-only replication phenotype of a molecular clone of simian immunodeficiency virus derived from the brain of a macaque with neuropathogenic infection. Virology. 2004;325:297–307. - PubMed
-
- Hahn BH. Shaw GM. deCock KM. Sharp PM. AIDS as a zoonosis: Scientific and public health implications. Science. 2000;287:607–614. - PubMed
-
- O'Neil SP. Suwyn C. Anderson DC. Niedziela G. Bradley J. Novembre FJ. Herndon JG. McClure HM. Correlation of acute humoral response with brain virus burden and survival time in pigtailed macaques infected with the neurovirulent simian immunodeficiency virus SIVsmmFGb. Am J Pathol. 2004;164:1157–1172. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

