Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response
- PMID: 20519338
- PMCID: PMC2905197
- DOI: 10.1093/jxb/erq154
Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response
Abstract
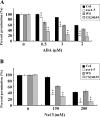
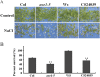
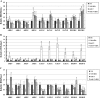
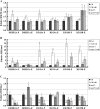
Histone modifications play an important role in the epigenetic regulation of gene expression. All histone modifications are reversible, which may therefore provide a flexible way for regulating gene expression during the plant's development and during the plant response to environmental stimuli. The reversible acetylation and deacetylation of specific lysine residues on core histones are catalysed by histone acetyltransferases and histone deacetylases (HDAs). HDA6 is an RPD3-type histone deacetylase in Arabidopsis. The Arabidopsis HDA6 mutant, axe1-5, and HDA6 RNA-interfering plants displayed a phenotype that was hypersensitive to ABA and salt stress. Compared with wild-type plants, the expression of the ABA and abiotic stress-responsive genes, ABI1, ABI2, KAT1, KAT2, DREB2A, RD29A, and RD29B, was decreased in axe1-5 and HDA6 RNA-interfering plants when treated with ABA or salt stress. It was found that both ABA and salt stress could enrich the gene activation markers, histone H3K9K14 acetylation, and H3K4 trimethylation, but decrease the gene repression marker, H3K9 dimethylation, of the ABA and abiotic stress-responsive genes. Our study indicates that HDA6-involved histone modifications modulate seed germination and the salt stress response, as well as ABA- and salt stress-induced gene expression in Arabidopsis.
Figures



References
-
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. - PubMed
-
- Bruce TJA, Matthes MC, Napier JA, Pickett JA. Stressful ‘memories’ of plants: evidence and possible mechanisms. Plant Science. 2007;173:603–608.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

