Accessory protein 5a is a major antagonist of the antiviral action of interferon against murine coronavirus
- PMID: 20519394
- PMCID: PMC2916514
- DOI: 10.1128/JVI.00385-10
Accessory protein 5a is a major antagonist of the antiviral action of interferon against murine coronavirus
Abstract
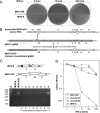
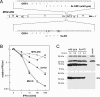
The type I interferon (IFN) response plays an essential role in the control of in vivo infection by the coronavirus mouse hepatitis virus (MHV). However, in vitro, most strains of MHV are largely resistant to the action of this cytokine, suggesting that MHV encodes one or more functions that antagonize or evade the IFN system. A particular strain of MHV, MHV-S, exhibited orders-of-magnitude higher sensitivity to IFN than prototype strain MHV-A59. Through construction of interstrain chimeric recombinants, the basis for the enhanced IFN sensitivity of MHV-S was found to map entirely to the region downstream of the spike gene, at the 3' end of the genome. Sequence analysis revealed that the major difference between the two strains in this region is the absence of gene 5a from MHV-S. Creation of a gene 5a knockout mutant of MHV-A59 demonstrated that a major component of IFN resistance maps to gene 5a. Conversely, insertion of gene 5a, or its homologs from related group 2 coronaviruses, at an upstream genomic position in an MHV-A59/S chimera restored IFN resistance. This is the first demonstration of a coronavirus gene product that can protect that same virus from the antiviral state induced by IFN. Neither protein kinase R, which phosphorylates eukaryotic initiation factor 2, nor oligoadenylate synthetase, which activates RNase L, was differentially activated in IFN-treated cells infected with MHV-A59 or MHV-S. Thus, the major IFN-induced antiviral activities that are specifically inhibited by MHV, and possibly by other coronaviruses, remain to be identified.
Figures

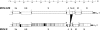

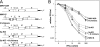

References
-
- Cervantes-Barragán, L., U. Kalinke, R. Züst, M. König, B. Reizis, C. López-Macías, V. Thiel, and B. Ludewig. 2009. Type I IFN-mediated protection of macrophages and dendritic cells secures control of murine coronavirus infection. J. Immunol. 182:1099-1106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

