Phosphorylation controls autoinhibition of cytoplasmic linker protein-170
- PMID: 20519438
- PMCID: PMC2912352
- DOI: 10.1091/mbc.e09-12-1036
Phosphorylation controls autoinhibition of cytoplasmic linker protein-170
Abstract
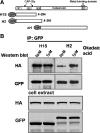
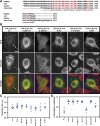
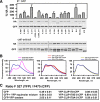
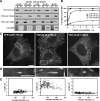
Cytoplasmic linker protein (CLIP)-170 is a microtubule (MT) plus-end-tracking protein that regulates MT dynamics and links MT plus ends to different intracellular structures. We have shown previously that intramolecular association between the N and C termini results in autoinhibition of CLIP-170, thus altering its binding to MTs and the dynactin subunit p150(Glued) (J. Cell Biol. 2004: 166, 1003-1014). In this study, we demonstrate that conformational changes in CLIP-170 are regulated by phosphorylation that enhances the affinity between the N- and C-terminal domains. By using site-directed mutagenesis and phosphoproteomic analysis, we mapped the phosphorylation sites in the third serine-rich region of CLIP-170. A phosphorylation-deficient mutant of CLIP-170 displays an "open" conformation and a higher binding affinity for growing MT ends and p150(Glued) as compared with nonmutated protein, whereas a phosphomimetic mutant confined to the "folded back" conformation shows decreased MT association and does not interact with p150(Glued). We conclude that phosphorylation regulates CLIP-170 conformational changes resulting in its autoinhibition.
Figures



References
-
- Akhmanova A., et al. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. - PubMed
-
- Akhmanova A., Steinmetz M. O. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 2008;9:309–322. - PubMed
-
- Arnal I., Heichette C., Diamantopoulos G. S., Chretien D. CLIP-170/tubulin-curved oligomers coassemble at microtubule ends and promote rescues. Curr. Biol. 2004;14:2086–2095. - PubMed
-
- Beausoleil S. A., Villen J., Gerber S. A., Rush J., Gygi S. P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006;24:1285–1292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

