Detection of large numbers of pneumococcal virulence genes in streptococci of the mitis group
- PMID: 20519466
- PMCID: PMC2916619
- DOI: 10.1128/JCM.01746-09
Detection of large numbers of pneumococcal virulence genes in streptococci of the mitis group
Abstract
Seven streptococcal isolates from the mitis group were analyzed for the presence of pneumococcal gene homologues by comparative genomic hybridization studies with microarrays based on open reading frames from the genomes of Streptococcus pneumoniae TIGR4 and R6. The diversity of pneumolysin (ply) and neuraminidase A (nanA) gene sequences was explored in more detail in a collection of 14 S. pseudopneumoniae and 29 mitis group isolates, respectively. The mitis group isolates used in the microarray experiments included a type strain (NCTC 12261), two S. mitis isolates from the nasopharynxes of children, one S. mitis isolate from a case of infective endocarditis, one S. mitis isolate from a dental abscess, and one S. oralis isolate and one S. pseudopneumoniae isolate from the nasopharynxes of children. The results of the microarray study showed that the 5 S. mitis isolates had homologues to between 67 and 82% of pneumococcal virulence genes, S. oralis hybridized to 83% of pneumococcal virulence genes, and S. pseudopneumoniae hybridized to 92% of identified pneumococcal virulence genes. Comparison of the pneumolysin, mitilysin (mly), and newly identified pseudopneumolysin (pply) gene sequences revealed that mly and pply genes are more closely related to each other than either is to ply. In contrast, the nanA gene sequences in the pneumococcus and streptococci from the mitis group are closely clustered together, sharing 99.4 to 99.7% sequence identity with pneumococcal nanA alleles.
Figures



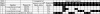
References
-
- Beighton, D., A. D. Carr, and B. A. Oppenheim. 1994. Identification of viridans streptococci associated with bacteraemia in neutropenic cancer patients. J. Med. Microbiol. 40:202-204. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

