Transcription of the transforming growth factor beta activating integrin beta8 subunit is regulated by SP3, AP-1, and the p38 pathway
- PMID: 20519498
- PMCID: PMC2915706
- DOI: 10.1074/jbc.M110.113977
Transcription of the transforming growth factor beta activating integrin beta8 subunit is regulated by SP3, AP-1, and the p38 pathway
Abstract
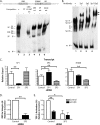
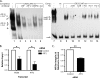
Integrin alphavbeta8 is a critical regulator of transforming growth factor beta activation in vasculogenesis during development, immune regulation, and endothelial/epithelial-mesenchymal homeostasis. Recent studies have suggested roles for integrin beta8 in the pathogenesis of chronic obstructive pulmonary disease, brain arteriovenous malformations, and select cancers (Araya, J., Cambier, S., Markovics, J. A., Wolters, P., Jablons, D., Hill, A., Finkbeiner, W., Jones, K., Broaddus, V. C., Sheppard, D., Barzcak, A., Xiao, Y., Erle, D. J., and Nishimura, S. L. (2007) J. Clin. Invest. 117, 3551-3562; Su, H., Kim, H., Pawlikowska, L., Kitamura, H., Shen, F., Cambier, S., Markovics, J., Lawton, M. T., Sidney, S., Bollen, A. W., Kwok, P. Y., Reichardt, L., Young, W. L., Yang, G. Y., and Nishimura, S. L. (2010) Am. J. Pathol. 176, 1018-1027; Culhane, A. C., and Quackenbush, J. (2009) Cancer Res. 69, 7480-7485; Cambier, S., Mu, D. Z., O'Connell, D., Boylen, K., Travis, W., Liu, W. H., Broaddus, V. C., and Nishimura, S. L. (2000) Cancer Res. 60, 7084-7093). Here we report the first identification and characterization of the promoter for ITGB8. We show that a SP binding site and a cyclic AMP response element (CRE) in the ITGB8 core promoter are required for its expression and that Sp1, Sp3, and several AP-1 transcription factors form a complex that binds to these sites in a p38-dependent manner. Furthermore, we demonstrate the requirement for Sp3, ATF-2, and p38 for the transcription and protein expression of integrin beta8. Additionally, reduction of SP3 or inhibition of p38 blocks alphavbeta8-mediated transforming growth factor beta activation. These results place integrin beta8 expression and activity under the control of ubiquitous transcription factors in a stress-activated and pro-inflammatory pathway.
Figures



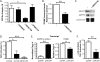

Similar articles
-
Interleukin-1beta induces increased transcriptional activation of the transforming growth factor-beta-activating integrin subunit beta8 through altering chromatin architecture.J Biol Chem. 2011 Oct 21;286(42):36864-74. doi: 10.1074/jbc.M111.276790. Epub 2011 Aug 30. J Biol Chem. 2011. PMID: 21878622 Free PMC article.
-
EGFR activation results in enhanced cyclooxygenase-2 expression through p38 mitogen-activated protein kinase-dependent activation of the Sp1/Sp3 transcription factors in human gliomas.Cancer Res. 2007 Jul 1;67(13):6121-9. doi: 10.1158/0008-5472.CAN-07-0141. Cancer Res. 2007. PMID: 17616668
-
Integrin alpha(v)beta8-mediated activation of transforming growth factor-beta by perivascular astrocytes: an angiogenic control switch.Am J Pathol. 2005 Jun;166(6):1883-94. doi: 10.1016/s0002-9440(10)62497-2. Am J Pathol. 2005. PMID: 15920172 Free PMC article.
-
Transcriptional regulation of the 5'-flanking region of the human transcription factor Sp3 gene by NF-1, c-Myb, B-Myb, AP-1 and E2F.Biochim Biophys Acta. 2008 May;1779(5):318-29. doi: 10.1016/j.bbagrm.2008.02.006. Epub 2008 Mar 10. Biochim Biophys Acta. 2008. PMID: 18342022
-
Laminin reduces expression of the human alpha6 integrin subunit gene by altering the level of the transcription factors Sp1 and Sp3.Invest Ophthalmol Vis Sci. 2007 Aug;48(8):3490-505. doi: 10.1167/iovs.07-0016. Invest Ophthalmol Vis Sci. 2007. PMID: 17652716
Cited by
-
Reprogramming tumor-infiltrating dendritic cells for CD103+ CD8+ mucosal T-cell differentiation and breast cancer rejection.Cancer Immunol Res. 2014 May;2(5):487-500. doi: 10.1158/2326-6066.CIR-13-0217. Epub 2014 Mar 4. Cancer Immunol Res. 2014. PMID: 24795361 Free PMC article.
-
MicroRNA-145 regulates human corneal epithelial differentiation.PLoS One. 2011;6(6):e21249. doi: 10.1371/journal.pone.0021249. Epub 2011 Jun 20. PLoS One. 2011. PMID: 21701675 Free PMC article.
-
A tumor-specific mechanism of Treg enrichment mediated by the integrin αvβ8.Sci Immunol. 2021 Mar 26;6(57):eabf0558. doi: 10.1126/sciimmunol.abf0558. Sci Immunol. 2021. PMID: 33771888 Free PMC article.
-
αvβ8 integrin adhesion and signaling pathways in development, physiology and disease.J Cell Sci. 2020 Jun 15;133(12):jcs239434. doi: 10.1242/jcs.239434. J Cell Sci. 2020. PMID: 32540905 Free PMC article. Review.
-
Paradoxical role of β8 integrin on angiogenesis and vasculogenic mimicry in glioblastoma.Cell Death Dis. 2022 Jun 8;13(6):536. doi: 10.1038/s41419-022-04959-7. Cell Death Dis. 2022. PMID: 35676251 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

