Serine residues in the cytosolic tail of the T-cell antigen receptor alpha-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein
- PMID: 20519503
- PMCID: PMC2911338
- DOI: 10.1074/jbc.M110.127936
Serine residues in the cytosolic tail of the T-cell antigen receptor alpha-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein
Abstract
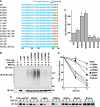
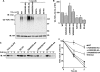
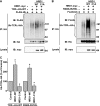
The T-cell antigen receptor (TCR) alpha-chain (TCRalpha) is a type I integral membrane protein that becomes ubiquitinated and targeted to the endoplasmic reticulum (ER)-associated degradation (ERAD) pathway when it fails to assemble into the heteromeric TCR complex. Remarkably, TCRalpha has a cytosolic tail of only five amino acid residues (i.e. RLWSS), none of which is the conventional ubiquitin acceptor, lysine. Herein we report that substitution of two conserved serine residues in the cytosolic tail of TCRalpha to alanine decreased ubiquitination, whereas placement of additional serine residues enhanced it. Moreover, replacement of the cytosolic serine residues by other ubiquitinatable residues (i.e. cysteine, threonine, or lysine) allowed ubiquitination to take place. Serine-dependent ubiquitination perfectly correlated with targeting of TCRalpha for ERAD. We also found that this ubiquitination was mediated by the ER-localized ubiquitin ligase, HRD1. These findings indicate that serine-dependent, HRD1-mediated ubiquitination targets TCRalpha to the ERAD pathway.
Figures



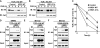

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

