An insight into the sialotranscriptome of Simulium nigrimanum, a black fly associated with fogo selvagem in South America
- PMID: 20519601
- PMCID: PMC2877412
- DOI: 10.4269/ajtmh.2010.09-0769
An insight into the sialotranscriptome of Simulium nigrimanum, a black fly associated with fogo selvagem in South America
Abstract
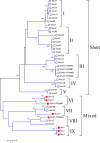
Pemphigus foliaceus is a life threatening skin disease that is associated with autoimmunity to desmoglein, a skin protein involved in the adhesion of keratinocytes. This disease is endemic in certain areas of South America, suggesting the mediation of environmental factors triggering autoimmunity. Among the possible environmental factors, exposure to bites of black flies, in particular Simulium nigrimanum has been suggested. In this work, we describe the sialotranscriptome of adult female S. nigrimanum flies. It reveals the complexity of the salivary potion of this insect, comprised by over 70 distinct genes within over 30 protein families, including several novel families, even when compared with the previously described sialotranscriptome of the autogenous black fly, S. vittatum. The uncovering of this sialotranscriptome provides a platform for testing pemphigus patient sera against recombinant salivary proteins from S. nigrimanum and for the discovery of novel pharmacologically active compounds.
Conflict of interest statement
Disclosure: Because JGV, VMP, and JMCR are government employees and this is a government work, the work is in the public domain in the United States. Notwithstanding any other agreements, the NIH reserves the right to provide the work to PubMedCentral for display and use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. You can establish rights outside of the U.S. subject to a government use license.
Figures


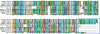

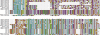
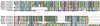

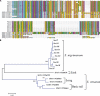
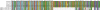
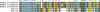

References
-
- Hilario-Vargas J, Dasher DA, Li N, Aoki V, Hans-Filho G, dos Santos V, Qaqish BF, Rivitti EA, Diaz LA. Prevalence of anti-desmoglein-3 antibodies in endemic regions of Fogo selvagem in Brazil. J Invest Dermatol. 2006;126:2044–2048. - PubMed
-
- Eaton DP, Diaz LA, Hans-Filho G, Santos VD, Aoki V, Friedman H, Rivitti EA, Sampaio SA, Gottlieb MS, Giudice GJ, Lopez A, Cupp EW. Comparison of black fly species (Diptera: Simuliidae) on an Amerindian reservation with a high prevalence of fogo selvagem to neighboring disease-free sites in the State of Mato Grosso do Sul, Brazil. The Cooperative Group on Fogo Selvagem Research. J Med Entomol. 1998;35:120–131. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

