Herbivore-induced SABATH methyltransferases of maize that methylate anthranilic acid using s-adenosyl-L-methionine
- PMID: 20519632
- PMCID: PMC2923889
- DOI: 10.1104/pp.110.158360
Herbivore-induced SABATH methyltransferases of maize that methylate anthranilic acid using s-adenosyl-L-methionine
Abstract
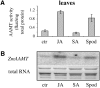
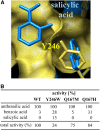
Volatile methyl esters are common constituents of plant volatiles with important functions in plant defense. To study the biosynthesis of these compounds, especially methyl anthranilate and methyl salicylate, we identified a group of methyltransferases that are members of the SABATH enzyme family in maize (Zea mays). In vitro biochemical characterization after bacterial expression revealed three S-adenosyl-L-methionine-dependent methyltransferases with high specificity for anthranilic acid as a substrate. Of these three proteins, Anthranilic Acid Methyltransferase1 (AAMT1) appears to be responsible for most of the S-adenosyl-L-methionine-dependent methyltransferase activity and methyl anthranilate formation observed in maize after herbivore damage. The enzymes may also be involved in the formation of low amounts of methyl salicylate, which are emitted from herbivore-damaged maize. Homology-based structural modeling combined with site-directed mutagenesis identified two amino acid residues, designated tyrosine-246 and glutamine-167 in AAMT1, which are responsible for the high specificity of AAMTs toward anthranilic acid. These residues are conserved in each of the three main clades of the SABATH family, indicating that the carboxyl methyltransferases are functionally separated by these clades. In maize, this gene family has diversified especially toward benzenoid carboxyl methyltransferases that accept anthranilic acid and benzoic acid.
Figures








References
-
- Barkman TJ, Martins TR, Sutton E, Stout JT. (2007) Positive selection for single amino acid change promotes substrate discrimination of a plant volatile-producing enzyme. Mol Biol Evol 24: 1320–1329 - PubMed
-
- Cardoza YJ, Tumlinson JH. (2006) Compatible and incompatible Xanthomonas infections differentially affect herbivore-induced volatile emission by pepper plants. J Chem Ecol 32: 1755–1768 - PubMed
-
- Chen F, D'Auria JC, Tholl D, Ross JR, Gershenzon J, Noel JP, Pichersky E. (2003) An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J 36: 577–588 - PubMed
-
- D'Alessandro M, Held M, Triponez Y, Turlings TCJ. (2006) The role of indole and other shikimic acid derived maize volatiles in the attraction of two parasitic wasps. J Chem Ecol 32: 2733–2748 - PubMed
-
- D'Auria JC, Chen F, Pichersky E. (2003) The SABATH family of MTS in Arabidopsis thaliana and other plant species. Romeo JT, , Integrative Phytochemistry: From Ethnobotany to Molecular Ecology. Recent Advances in Phytochemistry, Vol 37 Elsevier, Oxford, pp 253–283
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

