Differential replication of two chloroplast genome forms in heteroplasmic Chlamydomonas reinhardtii gametes contributes to alternative inheritance patterns
- PMID: 20519744
- PMCID: PMC2927747
- DOI: 10.1534/genetics.110.118265
Differential replication of two chloroplast genome forms in heteroplasmic Chlamydomonas reinhardtii gametes contributes to alternative inheritance patterns
Abstract
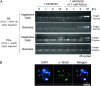
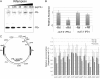
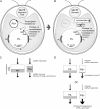
Two mechanisms for chloroplast DNA replication have been revealed through the study of an unusual heteroplasmic strain of the green alga Chlamydomonas reinhardtii. Heteroplasmy is a state in which more than one genome type occurs in a mitochondrion or chloroplast. The Chlamydomonas strain spa19 bears two distinct chloroplast genomes, termed PS+ and PS-. PS+ genomes predominate and are stably maintained in vegetative cells, despite their lack of known replication origins. In sexual crosses with spa19 as the mating type plus parent, however, PS+ genomes are transmitted in only approximately 25% of tetrads, whereas the PS- genomes are faithfully inherited in all progeny. In this research, we have explored the mechanism underlying this biased uniparental inheritance. We show that the relative reduction and dilution of PS+ vs. PS- genomes takes place during gametogenesis. Bromodeoxyuridine labeling, followed by immunoprecipitation and PCR, was used to compare replication activities of PS+ and PS- genomes. We found that the replication of PS+ genomes is specifically suppressed during gametogenesis and germination of zygospores, a phenomenon that also was observed when spa19 cells were treated with rifampicin, an inhibitor of the chloroplast RNA polymerase. Furthermore, when bromodeoxyuridine incorporation was compared at 11 sites within the chloroplast genome between vegetative cells, gametes, and rifampicin-treated cells by quantitative PCR, we found that incorporation was often reduced at the same sites in gametes that were also sensitive to rifampicin treatment. We conclude that a transcription-mediated form of DNA replication priming, which may be downregulated during gametogenesis, is indispensable for robust maintenance of PS+ genomes. These results highlight the potential for chloroplast genome copy number regulation through alternative replication strategies.
Figures




Similar articles
-
Uniparental inheritance of cpDNA and the genetic control of sexual differentiation in Chlamydomonas reinhardtii.J Plant Res. 2010 Mar;123(2):149-62. doi: 10.1007/s10265-009-0292-y. J Plant Res. 2010. PMID: 20196233 Review.
-
Sex-linked deubiquitinase establishes uniparental transmission of chloroplast DNA.Nat Commun. 2022 Mar 3;13(1):1133. doi: 10.1038/s41467-022-28807-6. Nat Commun. 2022. PMID: 35241655 Free PMC article.
-
Dynamic Changes in the Transcriptome and Methylome of Chlamydomonas reinhardtii throughout Its Life Cycle.Plant Physiol. 2015 Dec;169(4):2730-43. doi: 10.1104/pp.15.00861. Epub 2015 Oct 8. Plant Physiol. 2015. PMID: 26450704 Free PMC article.
-
A chloroplast-resident DNA methyltransferase is responsible for hypermethylation of chloroplast genes in Chlamydomonas maternal gametes.Proc Natl Acad Sci U S A. 2002 Apr 30;99(9):5925-30. doi: 10.1073/pnas.082120199. Proc Natl Acad Sci U S A. 2002. PMID: 11983892 Free PMC article.
-
The salvage/turnover/repair (STOR) model for uniparental inheritance in Chlamydomonas: DNA as a source of sustenance.J Hered. 1994 Sep-Oct;85(5):366-76. doi: 10.1093/oxfordjournals.jhered.a111481. J Hered. 1994. PMID: 7963452 Review.
Cited by
-
Chloroplast nucleoids as a transformable network revealed by live imaging with a microfluidic device.Commun Biol. 2018 May 17;1:47. doi: 10.1038/s42003-018-0055-1. eCollection 2018. Commun Biol. 2018. PMID: 30271930 Free PMC article.
-
Gsp1 triggers the sexual developmental program including inheritance of chloroplast DNA and mitochondrial DNA in Chlamydomonas reinhardtii.Plant Cell. 2012 Jun;24(6):2401-14. doi: 10.1105/tpc.112.097865. Epub 2012 Jun 19. Plant Cell. 2012. PMID: 22715041 Free PMC article.
-
Copper status of exposed microorganisms influences susceptibility to metallic nanoparticles.Environ Toxicol Chem. 2016 May;35(5):1148-58. doi: 10.1002/etc.3254. Epub 2016 Mar 9. Environ Toxicol Chem. 2016. PMID: 26387648 Free PMC article.
-
Why are most organelle genomes transmitted maternally?Bioessays. 2015 Jan;37(1):80-94. doi: 10.1002/bies.201400110. Epub 2014 Oct 10. Bioessays. 2015. PMID: 25302405 Free PMC article. Review.
-
Chloroplast competition is controlled by lipid biosynthesis in evening primroses.Proc Natl Acad Sci U S A. 2019 Mar 19;116(12):5665-5674. doi: 10.1073/pnas.1811661116. Epub 2019 Mar 4. Proc Natl Acad Sci U S A. 2019. PMID: 30833407 Free PMC article.
References
-
- Azhagiri, A. K., and P. Maliga, 2007. Exceptional paternal inheritance of plastids in Arabidopsis suggests that low-frequency leakage of plastids via pollen may be universal in plants. Plant J. 52 817–823. - PubMed
-
- Barr, C. M., M. Neiman and D. R. Taylor, 2005. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 168 39–50. - PubMed
-
- Bolen, P. L., N. W. Gillham and J. E. Boynton, 1980. Evidence for persistence of chloroplast markers in the heteroplasmic state in Chlamydomonas reinhardtii. Curr. Genet. 2 159–167. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

