Cooperation of DNA-PKcs and WRN helicase in the maintenance of telomeric D-loops
- PMID: 20519774
- PMCID: PMC2898018
- DOI: 10.18632/aging.100141
Cooperation of DNA-PKcs and WRN helicase in the maintenance of telomeric D-loops
Abstract
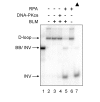
Werner syndrome is an inherited human progeriod syndrome caused by mutations in the gene encoding the Werner Syndrome protein, WRN. It has both 3'-5' DNA helicase and exonuclease activities, and is suggested to have roles in many aspects of DNA metabolism, including DNA repair and telomere maintenance. The DNA-PK complex also functions in both DNA double strand break repair and telomere maintenance. Interaction between WRN and the DNA-PK complex has been reported in DNA double strand break repair, but their possible cooperation at telomeres has not been reported. This study analyzes thein vitro and in vivo interaction at the telomere between WRN and DNA-PKcs, the catalytic subunit of DNA-PK. The results show that DNA-PKcs selectively stimulates WRN helicase but not WRN exonuclease in vitro, affecting that WRN helicase unwinds and promotes the release of the full-length invading strand of a telomere D-loop model substrate. In addition, the length of telomeric G-tails decreases in DNA-PKcs knockdown cells, and this phenotype is reversed by overexpression of WRN helicase. These results suggest that WRN and DNA-PKcs may cooperatively prevent G-tail shortening in vivo.
Conflict of interest statement
The authors of this manuscript have no conflict of interests to declare.
Figures






Comment in
-
New insight into telomere maintenance.Aging (Albany NY). 2010 May;2(5):255-6. doi: 10.18632/aging.100147. Aging (Albany NY). 2010. PMID: 20519779 Free PMC article. No abstract available.
-
Unraveling the roles of WRN and DNA-PKcs at telomeres.Aging (Albany NY). 2010 May;2(5):257-8. doi: 10.18632/aging.100150. Aging (Albany NY). 2010. PMID: 20519782 Free PMC article. No abstract available.
Similar articles
-
Unraveling the roles of WRN and DNA-PKcs at telomeres.Aging (Albany NY). 2010 May;2(5):257-8. doi: 10.18632/aging.100150. Aging (Albany NY). 2010. PMID: 20519782 Free PMC article. No abstract available.
-
Werner protein cooperates with the XRCC4-DNA ligase IV complex in end-processing.Biochemistry. 2008 Jul 15;47(28):7548-56. doi: 10.1021/bi702325t. Epub 2008 Jun 18. Biochemistry. 2008. PMID: 18558713 Free PMC article.
-
The Werner syndrome helicase/exonuclease processes mobile D-loops through branch migration and degradation.PLoS One. 2009;4(3):e4825. doi: 10.1371/journal.pone.0004825. Epub 2009 Mar 13. PLoS One. 2009. PMID: 19283071 Free PMC article.
-
Role of Werner syndrome gene product helicase in carcinogenesis and in resistance to genotoxins by cancer cells.Cancer Sci. 2008 May;99(5):843-8. doi: 10.1111/j.1349-7006.2008.00778.x. Epub 2008 Feb 26. Cancer Sci. 2008. PMID: 18312465 Free PMC article. Review.
-
Telomere ResQue and preservation--roles for the Werner syndrome protein and other RecQ helicases.Mech Ageing Dev. 2008 Jan-Feb;129(1-2):79-90. doi: 10.1016/j.mad.2007.10.007. Epub 2007 Oct 30. Mech Ageing Dev. 2008. PMID: 18054793 Review.
Cited by
-
Potential anti-aging agents suppress the level of constitutive mTOR- and DNA damage- signaling.Aging (Albany NY). 2012 Dec;4(12):952-65. doi: 10.18632/aging.100521. Aging (Albany NY). 2012. PMID: 23363784 Free PMC article.
-
RecQ Helicase Somatic Alterations in Cancer.Front Mol Biosci. 2022 Jun 15;9:887758. doi: 10.3389/fmolb.2022.887758. eCollection 2022. Front Mol Biosci. 2022. PMID: 35782872 Free PMC article. Review.
-
Telomerase-independent paths to immortality in predictable cancer subtypes.J Cancer. 2012;3:67-82. doi: 10.7150/jca.3965. Epub 2012 Jan 31. J Cancer. 2012. PMID: 22315652 Free PMC article.
-
Different non-synonymous polymorphisms modulate the interaction of the WRN protein to its protein partners and its enzymatic activities.Oncotarget. 2016 Dec 27;7(52):85680-85696. doi: 10.18632/oncotarget.13341. Oncotarget. 2016. PMID: 27863399 Free PMC article.
-
Unraveling the roles of WRN and DNA-PKcs at telomeres.Aging (Albany NY). 2010 May;2(5):257-8. doi: 10.18632/aging.100150. Aging (Albany NY). 2010. PMID: 20519782 Free PMC article. No abstract available.
References
-
- Martin GM. Genetics and aging; the Werner syndrome as a segmental progeroid syndrome. Adv Exp Med Biol. 1985;190:161–170. - PubMed
-
- Goto M. Werner's syndrome: from clinics to genetics. Clin Exp Rheumatol. 2000;18:760–766. - PubMed
-
- Shimamoto A, Sugimoto M, Furuichi Y. Molecular biology of Werner syndrome. Int J Clin Oncol. 2004;9:288–298. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

