Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation
- PMID: 20520723
- PMCID: PMC2875390
- DOI: 10.1371/journal.pone.0010790
Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation
Abstract
Background: The accessory gene regulator (agr) and staphylococcal accessory regulator (sarA) play opposing roles in Staphylococcus aureus biofilm formation. There is mounting evidence to suggest that these opposing roles are therapeutically relevant in that mutation of agr results in increased biofilm formation and decreased antibiotic susceptibility while mutation of sarA has the opposite effect. To the extent that induction of agr or inhibition of sarA could potentially be used to limit biofilm formation, this makes it important to understand the epistatic relationships between these two loci.
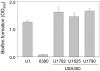
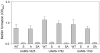
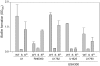
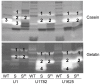
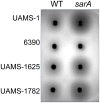
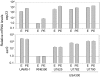
Methodology/principal findings: We generated isogenic sarA and agr mutants in clinical isolates of S. aureus and assessed the relative impact on biofilm formation. Mutation of agr resulted in an increased capacity to form a biofilm in the 8325-4 laboratory strain RN6390 but had little impact in clinical isolates S. aureus. In contrast, mutation of sarA resulted in a reduced capacity to form a biofilm in all clinical isolates irrespective of the functional status of agr. This suggests that the regulatory role of sarA in biofilm formation is independent of the interaction between sarA and agr and that sarA is epistatic to agr in this context. This was confirmed by demonstrating that restoration of sarA function restored the ability to form a biofilm even in the corresponding agr mutants. Mutation of sarA in clinical isolates also resulted in increased production of extracellular proteases and extracellular nucleases, both of which contributed to the biofilm-deficient phenotype of sarA mutants. However, studies comparing different strains with and without proteases inhibitors and/or mutation of the nuclease genes demonstrated that the agr-independent, sarA-mediated repression of extracellular proteases plays a primary role in this regard.
Conclusions and significance: The results we report suggest that inhibitors of sarA-mediated regulation could be used to limit biofilm formation in S. aureus and that the efficacy of such inhibitors would not be limited by spontaneous mutation of agr in the human host.
Conflict of interest statement
Figures
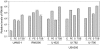




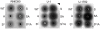



References
-
- Brady RA, Leid JG, Calhoun JH, Costerton JW, Shirtliff ME. Osteomyelitis and the role of biofilms in chronic infection. FEMS Immunol. Med Microbiol. 2008;52:13–22. - PubMed
-
- O'Gara JP. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett. 2007;270:179–188. - PubMed
-
- Coelho LR, Souza RR, Ferreira FA, Guimarães MA, Ferreira-Carvalho BT, et al. agr RNAIII divergently regulates glucose-induced biofilm formation in clinical isolates of Staphylococcus aureus. Microbiology. 2008;154:3480–3490. - PubMed
-
- Kong KF, Vuong C, Otto M. Staphylococcus quorum sensing in biofilm formation and infection. Int J Med Microbiol. 2006;296:133–139. - PubMed
-
- Vuong C, Saenz HL, Götz F, Otto M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis. 2000;182:1688–1693. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

