Active chromatin marks are retained on X chromosomes lacking gene or repeat silencing despite XIST/Xist expression in somatic cell hybrids
- PMID: 20520737
- PMCID: PMC2875404
- DOI: 10.1371/journal.pone.0010787
Active chromatin marks are retained on X chromosomes lacking gene or repeat silencing despite XIST/Xist expression in somatic cell hybrids
Abstract
Background: X-chromosome inactivation occurs early in mammalian development and results in the inactive X chromosome acquiring numerous hallmarks of heterochromatin. While XIST is a key player in the inactivation process, the method of action of this ncRNA is yet to be determined.
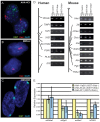
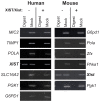
Methodology/principal findings: To assess which features of heterochromatin may be directly recruited by the expression and localization of the XIST RNA we have analyzed a mouse/human somatic cell hybrid in which expression of human and mouse XIST/Xist has been induced from the active X by demethylation. Such hybrids had previously been demonstrated to disconnect XIST/Xist expression from gene silencing and we confirm maintenance of X-linked gene expression, even close to the Xist locus, despite the localized expression of mouse Xist.
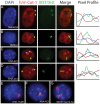
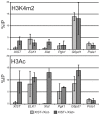
Conclusions/significance: Loss of the active chromatin marks H3 acetylation and H3 lysine 4 methylation was not observed upon XIST/Xist expression, nor was there a gain of DNA methylation; thus these marks of facultative heterochromatin are not solely dependent upon Xist expression. Cot-1 holes, regions of depleted RNA hybridization with a Cot-1 probe, were observed upon Xist expression; however, these were at reduced frequency and intensity in these somatic cells. Domains of human Cot-1 transcription were observed corresponding to the human chromosomes in the somatic cell hybrids. The Cot-1 domain of the X was not reduced with the expression of XIST, which fails to localize to the human X chromosome in a mouse somatic cell background. The human inactive X in a mouse/human hybrid cell also shows delocalized XIST expression and an ongoing Cot-1 domain, despite X-linked gene silencing. These results are consistent with recent reports separating Cot-1 silencing from genic silencing, but also demonstrate repetitive element expression from an otherwise silent X chromosome in these hybrid cells.
Conflict of interest statement
Figures
References
-
- Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. - PubMed
-
- Huynh KD, Lee JT. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature. 2003;426:857–862. - PubMed
-
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

