Thiazolyl peptide antibiotic biosynthesis: a cascade of post-translational modifications on ribosomal nascent proteins
- PMID: 20522549
- PMCID: PMC2934618
- DOI: 10.1074/jbc.R110.135970
Thiazolyl peptide antibiotic biosynthesis: a cascade of post-translational modifications on ribosomal nascent proteins
Abstract
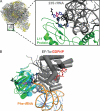
Antibiotics of the thiocillin, GE2270A, and thiostrepton class, which block steps in bacterial protein synthesis, contain a trithiazolyl (tetrahydro)pyridine core that provides the architectural constraints for high affinity binding to either the 50 S ribosomal subunit or elongation factor Tu. These mature antibiotic scaffolds arise from a cascade of post-translational modifications on 50-60-residue prepeptide precursors that trim away the N-terminal leader sequences (approximately 40 residues) while the C-terminal 14-18 residues are converted into the mature scaffold. In the producing microbes, the genes encoding the prepeptide open reading frames are flanked in biosynthetic clusters by genes encoding post-translational modification enzymes that carry out lantibiotic-type dehydrations of Ser and Thr residues to dehydroamino acid side chains, cyclodehydration and oxidation of cysteines to thiazoles, and condensation of two dehydroalanine residues en route to the (tetrahydro)pyridine core. The trithiazolyl pyridine framework thus arises from post-translational modification of the peptide backbone of three Cys and two Ser residues of the prepeptide.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

