On inferring presence of an individual in a mixture: a Bayesian approach
- PMID: 20522729
- PMCID: PMC2950790
- DOI: 10.1093/biostatistics/kxq035
On inferring presence of an individual in a mixture: a Bayesian approach
Abstract
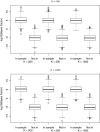
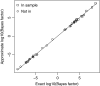
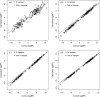
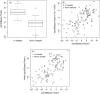
Homer and others (2008. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genetics 4, e1000167) recently showed that, given allele frequency data for a large number of single nucleotide polymorphisms in a sample together with corresponding population "reference" frequencies, by typing an individual's DNA sample at the same set of loci it can be inferred whether or not the individual was a member of the sample. This observation has been responsible for precautionary removal of large amounts of summary data from public access. This and further work on the problem has followed a frequentist approach. This paper sets out a Bayesian analysis of this problem which clarifies the role of the reference frequencies and allows incorporation of prior probabilities of the individual's membership in the sample.
Figures
Similar articles
-
Scalable linkage-disequilibrium-based selective sweep detection: a performance guide.Gigascience. 2016 Feb 8;5:7. doi: 10.1186/s13742-016-0114-9. eCollection 2016. Gigascience. 2016. PMID: 26862394 Free PMC article.
-
Inferring the history of population size change from genome-wide SNP data.Mol Biol Evol. 2012 Dec;29(12):3653-67. doi: 10.1093/molbev/mss175. Epub 2012 Jul 10. Mol Biol Evol. 2012. PMID: 22787284
-
Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays.PLoS Genet. 2008 Aug 29;4(8):e1000167. doi: 10.1371/journal.pgen.1000167. PLoS Genet. 2008. PMID: 18769715 Free PMC article.
-
[Analysis and application of haplotype in forensic medicine].Fa Yi Xue Za Zhi. 2009 Apr;25(2):133-7. Fa Yi Xue Za Zhi. 2009. PMID: 19537256 Review. Chinese.
-
On the use of kernel approximate Bayesian computation to infer population history.Genes Genet Syst. 2015;90(3):153-62. doi: 10.1266/ggs.90.153. Genes Genet Syst. 2015. PMID: 26510570 Review.
Cited by
-
Genome Reconstruction Attacks Against Genomic Data-Sharing Beacons.Proc Priv Enhanc Technol. 2021;2021(3):28-48. doi: 10.2478/popets-2021-0036. Epub 2021 Apr 26. Proc Priv Enhanc Technol. 2021. PMID: 34746296 Free PMC article.
-
Beacon Reconstruction Attack: Reconstruction of genomes in genomic data-sharing beacons using summary statistics.Bioinformatics. 2025 Jun 2;41(6):btaf273. doi: 10.1093/bioinformatics/btaf273. Bioinformatics. 2025. PMID: 40388204 Free PMC article.
-
Assessing and managing risk when sharing aggregate genetic variant data.Nat Rev Genet. 2011 Sep 16;12(10):730-6. doi: 10.1038/nrg3067. Nat Rev Genet. 2011. PMID: 21921928 Free PMC article. Review.
-
Identifiability in biobanks: models, measures, and mitigation strategies.Hum Genet. 2011 Sep;130(3):383-92. doi: 10.1007/s00439-011-1042-5. Epub 2011 Jul 8. Hum Genet. 2011. PMID: 21739176 Free PMC article. Review.
-
Participant identification in genetic association studies: improved methods and practical implications.Int J Epidemiol. 2011 Dec;40(6):1629-42. doi: 10.1093/ije/dyr149. Int J Epidemiol. 2011. PMID: 22158671 Free PMC article.
References
-
- Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Annals of Statistics. 2003;32:407–499.
-
- Heath SC, Gut IG, Brennan P, McKay JD, Bencko V, Fabianova E, Foretova L, Georges M, Janout V, Kabesch M. Investigation of the fine structure of European populations with applications to disease association studies. European Journal of Human Genetics. 2008;16:1413–1429. and others. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

