Genome-wide identification of calcium-response factor (CaRF) binding sites predicts a role in regulation of neuronal signaling pathways
- PMID: 20523734
- PMCID: PMC2877716
- DOI: 10.1371/journal.pone.0010870
Genome-wide identification of calcium-response factor (CaRF) binding sites predicts a role in regulation of neuronal signaling pathways
Abstract
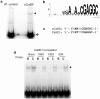
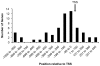
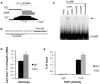
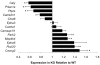
Calcium-Response Factor (CaRF) was first identified as a transcription factor based on its affinity for a neuronal-selective calcium-response element (CaRE1) in the gene encoding Brain-Derived Neurotrophic Factor (BDNF). However, because CaRF shares no homology with other transcription factors, its properties and gene targets have remained unknown. Here we show that the DNA binding domain of CaRF has been highly conserved across evolution and that CaRF binds DNA directly in a sequence-specific manner in the absence of other eukaryotic cofactors. Using a binding site selection screen we identify a high-affinity consensus CaRF response element (cCaRE) that shares significant homology with the CaRE1 element of Bdnf. In a genome-wide chromatin immunoprecipitation analysis (ChIP-Seq), we identified 176 sites of CaRF-specific binding (peaks) in neuronal genomic DNA. 128 of these peaks are within 10kB of an annotated gene, and 60 are within 1kB of an annotated transcriptional start site. At least 138 of the CaRF peaks contain a common 10-bp motif with strong statistical similarity to the cCaRE, and we provide evidence predicting that CaRF can bind independently to at least 64.5% of these motifs in vitro. Analysis of this set of putative CaRF targets suggests the enrichment of genes that regulate intracellular signaling cascades. Finally we demonstrate that expression of a subset of these target genes is altered in the cortex of Carf knockout (KO) mice. Together these data strongly support the characterization of CaRF as a unique transcription factor and provide the first insight into the program of CaRF-regulated transcription in neurons.
Conflict of interest statement
Figures



References
-
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10:252–263. - PubMed
-
- Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. - PubMed
-
- Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

