The use of orthologous sequences to predict the impact of amino acid substitutions on protein function
- PMID: 20523748
- PMCID: PMC2877731
- DOI: 10.1371/journal.pgen.1000968
The use of orthologous sequences to predict the impact of amino acid substitutions on protein function
Abstract
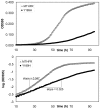
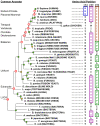
Computational predictions of the functional impact of genetic variation play a critical role in human genetics research. For nonsynonymous coding variants, most prediction algorithms make use of patterns of amino acid substitutions observed among homologous proteins at a given site. In particular, substitutions observed in orthologous proteins from other species are often assumed to be tolerated in the human protein as well. We examined this assumption by evaluating a panel of nonsynonymous mutants of a prototypical human enzyme, methylenetetrahydrofolate reductase (MTHFR), in a yeast cell-based functional assay. As expected, substitutions in human MTHFR at sites that are well-conserved across distant orthologs result in an impaired enzyme, while substitutions present in recently diverged sequences (including a 9-site mutant that "resurrects" the human-macaque ancestor) result in a functional enzyme. We also interrogated 30 sites with varying degrees of conservation by creating substitutions in the human enzyme that are accepted in at least one ortholog of MTHFR. Quite surprisingly, most of these substitutions were deleterious to the human enzyme. The results suggest that selective constraints vary between phylogenetic lineages such that inclusion of distant orthologs to infer selective pressures on the human enzyme may be misleading. We propose that homologous proteins are best used to reconstruct ancestral sequences and infer amino acid conservation among only direct lineal ancestors of a particular protein. We show that such an "ancestral site preservation" measure outperforms other prediction methods, not only in our selected set for MTHFR, but also in an exhaustive set of E. coli LacI mutants.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. - PubMed
-
- Miller MP, Kumar S. Understanding human disease mutations through the use of interspecific genetic variation. Hum Mol Genet. 2001;10:2319–2328. - PubMed
-
- Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

