Global migration dynamics underlie evolution and persistence of human influenza A (H3N2)
- PMID: 20523898
- PMCID: PMC2877742
- DOI: 10.1371/journal.ppat.1000918
Global migration dynamics underlie evolution and persistence of human influenza A (H3N2)
Abstract
The global migration patterns of influenza viruses have profound implications for the evolutionary and epidemiological dynamics of the disease. We developed a novel approach to reconstruct the genetic history of human influenza A (H3N2) collected worldwide over 1998 to 2009 and used it to infer the global network of influenza transmission. Consistent with previous models, we find that China and Southeast Asia lie at the center of this global network. However, we also find that strains of influenza circulate outside of Asia for multiple seasons, persisting through dynamic migration between northern and southern regions. The USA acts as the primary hub of temperate transmission and, together with China and Southeast Asia, forms the trunk of influenza's evolutionary tree. These findings suggest that antiviral use outside of China and Southeast Asia may lead to the evolution of long-term local and potentially global antiviral resistance. Our results might also aid the design of surveillance efforts and of vaccines better tailored to different geographic regions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
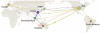

 -axis. Tracing a vertical line gives a contemporaneous cross-section of virus isolates. The genealogy is sorted so that lineages that leave more descendants are placed higher on the
-axis. Tracing a vertical line gives a contemporaneous cross-section of virus isolates. The genealogy is sorted so that lineages that leave more descendants are placed higher on the  -axis than other, less successful lineages. This sorting places the trunk along a rough diagonal, and it places lineages that are more genetically similar to the trunk higher on the
-axis than other, less successful lineages. This sorting places the trunk along a rough diagonal, and it places lineages that are more genetically similar to the trunk higher on the  -axis than lineages that are farther away from the trunk. The tree shown is the highest posterior tree generated by the Markov chain Monte Carlo (MCMC) procedure implemented in the software program Migrate v3.0.8 , .
-axis than lineages that are farther away from the trunk. The tree shown is the highest posterior tree generated by the Markov chain Monte Carlo (MCMC) procedure implemented in the software program Migrate v3.0.8 , .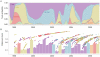
 -axis, we can be fairly certain that the trunk of the genealogy is in this location. Other times, when there is a mix of colors, we are not so certain. (B) Distance to the trunk, measured in terms of years, for each sampled influenza sequence. Here, points represent individual tips of the influenza tree colored as in Figure 2. The height of each point on the
-axis, we can be fairly certain that the trunk of the genealogy is in this location. Other times, when there is a mix of colors, we are not so certain. (B) Distance to the trunk, measured in terms of years, for each sampled influenza sequence. Here, points represent individual tips of the influenza tree colored as in Figure 2. The height of each point on the  -axis shows the mean distance to the trunk across the full range of estimated genealogies. Bars identify the closest sample to the trunk within a 4 month window of time. Bars are colored according to regions of these samples.
-axis shows the mean distance to the trunk across the full range of estimated genealogies. Bars identify the closest sample to the trunk within a 4 month window of time. Bars are colored according to regions of these samples.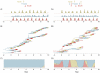
 -axis, we can be fairly certain that the trunk of the genealogy is in this location. Other times, when there is a mix of colors, we are not so certain.
-axis, we can be fairly certain that the trunk of the genealogy is in this location. Other times, when there is a mix of colors, we are not so certain.References
-
- World Health Organization. Fact sheet Number 211. 2009. Influenza. URL http://www.who.int/mediacentre/factsheets/fs211/en/
-
- Russell CA, Jones TC, Barr IG, Cox NJ, Garten RJ, et al. The global circulation of seasonal influenza A (H3N2) viruses. Science. 2008;320:340–346. - PubMed
-
- Ferguson NM, Galvani AP, Bush RM. Ecological and immunological determinants of influenza evolution. Nature. 2003;422:428–433. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

