The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin
- PMID: 20523900
- PMCID: PMC2877744
- DOI: 10.1371/journal.ppat.1000919
The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin
Abstract
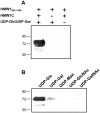
The Haemophilus influenzae HMW1 adhesin is a high-molecular weight protein that is secreted by the bacterial two-partner secretion pathway and mediates adherence to respiratory epithelium, an essential early step in the pathogenesis of H. influenzae disease. In recent work, we discovered that HMW1 is a glycoprotein and undergoes N-linked glycosylation at multiple asparagine residues with simple hexose units rather than N-acetylated hexose units, revealing an unusual N-glycosidic linkage and suggesting a new glycosyltransferase activity. Glycosylation protects HMW1 against premature degradation during the process of secretion and facilitates HMW1 tethering to the bacterial surface, a prerequisite for HMW1-mediated adherence. In the current study, we establish that the enzyme responsible for glycosylation of HMW1 is a protein called HMW1C, which is encoded by the hmw1 gene cluster and shares homology with a group of bacterial proteins that are generally associated with two-partner secretion systems. In addition, we demonstrate that HMW1C is capable of transferring glucose and galactose to HMW1 and is also able to generate hexose-hexose bonds. Our results define a new family of bacterial glycosyltransferases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The Actinobacillus pleuropneumoniae HMW1C-like glycosyltransferase mediates N-linked glycosylation of the Haemophilus influenzae HMW1 adhesin.PLoS One. 2010 Dec 30;5(12):e15888. doi: 10.1371/journal.pone.0015888. PLoS One. 2010. PMID: 21209858 Free PMC article.
-
The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis.Mol Microbiol. 2003 May;48(3):737-51. doi: 10.1046/j.1365-2958.2003.03450.x. Mol Microbiol. 2003. PMID: 12694618
-
Unconventional N-Linked Glycosylation Promotes Trimeric Autotransporter Function in Kingella kingae and Aggregatibacter aphrophilus.mBio. 2015 Aug 25;6(4):e01206-15. doi: 10.1128/mBio.01206-15. mBio. 2015. PMID: 26307167 Free PMC article.
-
A prototype two-partner secretion pathway: the Haemophilus influenzae HMW1 and HMW2 adhesin systems.Trends Microbiol. 2009 Aug;17(8):355-60. doi: 10.1016/j.tim.2009.06.002. Epub 2009 Aug 5. Trends Microbiol. 2009. PMID: 19660953 Review.
-
Glycosylation and biogenesis of a family of serine-rich bacterial adhesins.Microbiology (Reading). 2009 Feb;155(Pt 2):317-327. doi: 10.1099/mic.0.025221-0. Microbiology (Reading). 2009. PMID: 19202081 Review.
Cited by
-
A two-step enzymatic glycosylation of polypeptides with complex N-glycans.Bioorg Med Chem. 2013 Apr 15;21(8):2262-2270. doi: 10.1016/j.bmc.2013.02.007. Epub 2013 Feb 18. Bioorg Med Chem. 2013. PMID: 23477942 Free PMC article.
-
Cytoplasmic N-glycosyltransferase of Actinobacillus pleuropneumoniae is an inverting enzyme and recognizes the NX(S/T) consensus sequence.J Biol Chem. 2011 Oct 7;286(40):35267-74. doi: 10.1074/jbc.M111.277160. Epub 2011 Aug 18. J Biol Chem. 2011. PMID: 21852240 Free PMC article.
-
Spindly is a nucleocytosolic O-fucosyltransferase in Dictyostelium and related proteins are widespread in protists and bacteria.Glycobiology. 2023 Apr 19;33(3):225-244. doi: 10.1093/glycob/cwac071. Glycobiology. 2023. PMID: 36250576 Free PMC article.
-
Leloir Glycosyltransferases in Applied Biocatalysis: A Multidisciplinary Approach.Int J Mol Sci. 2019 Oct 23;20(21):5263. doi: 10.3390/ijms20215263. Int J Mol Sci. 2019. PMID: 31652818 Free PMC article. Review.
-
Differential recognition of Haemophilus influenzae whole bacterial cells and isolated lipooligosaccharides by galactose-specific lectins.Sci Rep. 2018 Nov 2;8(1):16292. doi: 10.1038/s41598-018-34383-x. Sci Rep. 2018. PMID: 30389954 Free PMC article.
References
-
- Benz I, Schmidt MA. Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA-1 adhesin. Mol Microbiol. 2001;40:1403–1413. - PubMed
-
- Castric P. pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology. 1995;141:1247–1254. - PubMed
-
- Doig P, Kinsella N, Guerry P, Trust TJ. Characterization of a post-translational modification of Campylobacter flagellin: identification of a sero-specific glycosyl moiety. Mol Microbiol. 1996;19:379–387. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

