Epistasis of transcriptomes reveals synergism between transcriptional activators Hnf1alpha and Hnf4alpha
- PMID: 20523905
- PMCID: PMC2877749
- DOI: 10.1371/journal.pgen.1000970
Epistasis of transcriptomes reveals synergism between transcriptional activators Hnf1alpha and Hnf4alpha
Abstract
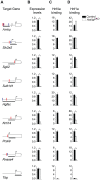
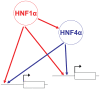
The transcription of individual genes is determined by combinatorial interactions between DNA-binding transcription factors. The current challenge is to understand how such combinatorial interactions regulate broad genetic programs that underlie cellular functions and disease. The transcription factors Hnf1alpha and Hnf4alpha control pancreatic islet beta-cell function and growth, and mutations in their genes cause closely related forms of diabetes. We have now exploited genetic epistasis to examine how Hnf1alpha and Hnf4alpha functionally interact in pancreatic islets. Expression profiling in islets from either Hnf1a(+/-) or pancreas-specific Hnf4a mutant mice showed that the two transcription factors regulate a strikingly similar set of genes. We integrated expression and genomic binding studies and show that the shared transcriptional phenotype of these two mutant models is linked to common direct targets, rather than to known effects of Hnf1alpha on Hnf4a gene transcription. Epistasis analysis with transcriptomes of single- and double-mutant islets revealed that Hnf1alpha and Hnf4alpha regulate common targets synergistically. Hnf1alpha binding in Hnf4a-deficient islets was decreased in selected targets, but remained unaltered in others, thus suggesting that the mechanisms for synergistic regulation are gene-specific. These findings provide an in vivo strategy to study combinatorial gene regulation and reveal how Hnf1alpha and Hnf4alpha control a common islet-cell regulatory program that is defective in human monogenic diabetes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
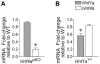



References
-
- Carey M, Smale ST. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. Transcriptional regulation in eukaryotes concepts, strategies, and techniques.
-
- Du W, Thanos D, Maniatis T. Mechanisms of transcriptional synergism between distinct virus-inducible enhancer elements. Cell. 1993;74:887–898. - PubMed
-
- Yuh CH, Bolouri H, Davidson EH. Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

