Aging and chronic sun exposure cause distinct epigenetic changes in human skin
- PMID: 20523906
- PMCID: PMC2877750
- DOI: 10.1371/journal.pgen.1000971
Aging and chronic sun exposure cause distinct epigenetic changes in human skin
Abstract
Epigenetic changes are widely considered to play an important role in aging, but experimental evidence to support this hypothesis has been scarce. We have used array-based analysis to determine genome-scale DNA methylation patterns from human skin samples and to investigate the effects of aging, chronic sun exposure, and tissue variation. Our results reveal a high degree of tissue specificity in the methylation patterns and also showed very little interindividual variation within tissues. Data stratification by age revealed that DNA from older individuals was characterized by a specific hypermethylation pattern affecting less than 1% of the markers analyzed. Interestingly, stratification by sun exposure produced a fundamentally different pattern with a significant trend towards hypomethylation. Our results thus identify defined age-related DNA methylation changes and suggest that these alterations might contribute to the phenotypic changes associated with skin aging.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

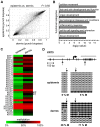
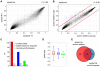
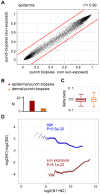
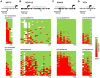
References
-
- Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. - PubMed
-
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. - PubMed
-
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

