Elucidating the tertiary structure of protein ions in vacuo with site specific photoinitiated radical reactions
- PMID: 20524634
- PMCID: PMC2907658
- DOI: 10.1021/ja910665d
Elucidating the tertiary structure of protein ions in vacuo with site specific photoinitiated radical reactions
Abstract
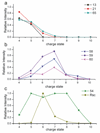
A new method for identifying residue specific through space contacts as a function of protein secondary and tertiary structure in the gas phase is presented. Photodissociation of a non-native carbon-iodine bond incorporated into Tyr59 of ubiquitin yields a radical site specifically at that residue. The subsequent radical migration is shown to be highly dependent on the structure of the protein. Radical-directed dissociation (RDD) of low charge states, which adopt compact structures, generates backbone fragmentation that is prominently distributed throughout the protein sequence, including residues that are distant in sequence from Tyr59. Higher charge states of ubiquitin, which adopt elongated, unfolded structures, yield RDD that is primarily nearby in sequence to Tyr59. Regardless of which structure is probed, information at the residue-level is obtained by examining specific radical-donor and radical-acceptor pairs. The relative importance of a particular interaction pair for a specific conformation can be revealed by tracking the charge state dependence of the dissociation. Structurally important contact pairs exhibit strong and concerted changes in relative intensities as a function of charge state and can also be used to reveal structural features which persist among different protein structures. Moreover, incorporation of distance constraint information into molecular mechanics conformational searches can be used to drive the search toward relevant conformational space. Implementation of this approach has revealed highly stable, previously undiscovered structures for the +4 and +6 charge states of ubiquitin, which bear little resemblance to the crystal structure.
Figures




Similar articles
-
Bond-specific dissociation following excitation energy transfer for distance constraint determination in the gas phase.J Am Chem Soc. 2014 Sep 24;136(38):13363-70. doi: 10.1021/ja507215q. Epub 2014 Sep 11. J Am Chem Soc. 2014. PMID: 25174489 Free PMC article.
-
TEMPO-Assisted Free-Radical-Initiated Peptide Sequencing Mass Spectrometry for Ubiquitin Ions: An Insight on the Gas-Phase Conformations.J Am Soc Mass Spectrom. 2022 Mar 2;33(3):471-481. doi: 10.1021/jasms.1c00313. Epub 2022 Jan 31. J Am Soc Mass Spectrom. 2022. PMID: 35099967
-
Detailed unfolding and folding of gaseous ubiquitin ions characterized by electron capture dissociation.J Am Chem Soc. 2002 Jun 5;124(22):6407-20. doi: 10.1021/ja012267j. J Am Chem Soc. 2002. PMID: 12033872
-
Radical-driven peptide backbone dissociation tandem mass spectrometry.Mass Spectrom Rev. 2015 Mar-Apr;34(2):116-32. doi: 10.1002/mas.21426. Epub 2014 May 26. Mass Spectrom Rev. 2015. PMID: 24863492 Review.
-
Mass Spectrometry-Based Fast Photochemical Oxidation of Proteins (FPOP) for Higher Order Structure Characterization.Acc Chem Res. 2018 Mar 20;51(3):736-744. doi: 10.1021/acs.accounts.7b00593. Epub 2018 Feb 16. Acc Chem Res. 2018. PMID: 29450991 Free PMC article. Review.
Cited by
-
Changes in protein structure monitored by use of gas-phase hydrogen/deuterium exchange.Proteomics. 2015 Aug;15(16):2842-50. doi: 10.1002/pmic.201400440. Epub 2015 Mar 18. Proteomics. 2015. PMID: 25603979 Free PMC article.
-
Computational Insights into Compaction of Gas-Phase Protein and Protein Complex Ions in Native Ion Mobility-Mass Spectrometry.Trends Analyt Chem. 2019 Jul;116:282-291. doi: 10.1016/j.trac.2019.04.023. Epub 2019 Apr 30. Trends Analyt Chem. 2019. PMID: 31983791 Free PMC article.
-
Enhancing Biomolecule Analysis and 2DMS Experiments by Implementation of (Activated Ion) 193 nm UVPD on a FT-ICR Mass Spectrometer.Anal Chem. 2022 Nov 15;94(45):15631-15638. doi: 10.1021/acs.analchem.2c02354. Epub 2022 Nov 1. Anal Chem. 2022. PMID: 36317856 Free PMC article.
-
Bond-specific dissociation following excitation energy transfer for distance constraint determination in the gas phase.J Am Chem Soc. 2014 Sep 24;136(38):13363-70. doi: 10.1021/ja507215q. Epub 2014 Sep 11. J Am Chem Soc. 2014. PMID: 25174489 Free PMC article.
-
Charge transfer dissociation (CTD) mass spectrometry of peptide cations using kiloelectronvolt helium cations.J Am Soc Mass Spectrom. 2014 Nov;25(11):1939-43. doi: 10.1007/s13361-014-0989-6. Epub 2014 Sep 18. J Am Soc Mass Spectrom. 2014. PMID: 25231159
References
-
- Valentine SJ, Counterman AE, Clemmer DE. J. Am. Soc. Mass Spectrom. 1997;8:954–961. - PubMed
-
- Shelimov KB, Clemmer DE, Hudgins RR, Jarrold MF. J. Am. Chem. Soc. 1997;119:2240–2248.
-
- Clemmer DE, Hudgins RR, Jarrold MF. J. Am. Chem. Soc. 1995;117:10141–10142.
-
- Hoaglund CS, Valentine SJ, Sporleder CR, Reilly JP, Clemmer DE. Anal. Chem. 1998;70:2236–2242. - PubMed
-
- Purves RW, Barnett DA, Ells B, Guevremont R. J. Am. Soc. Mass Spectrom. 2000;11:738–745. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

