Integration of transcriptional inputs at promoters of the arabinose catabolic pathway
- PMID: 20525212
- PMCID: PMC2893085
- DOI: 10.1186/1752-0509-4-75
Integration of transcriptional inputs at promoters of the arabinose catabolic pathway
Abstract
Background: Most modelling efforts of transcriptional networks involve estimations of in vivo concentrations of components, binding affinities and reaction rates, derived from in vitro biochemical assays. These assays are difficult and in vitro measurements may not approximate actual in vivo conditions. Alternatively, changes in transcription factor activity can be estimated by using partially specified models which estimate the "hidden functions" of transcription factor concentration changes; however, non-unique solutions are a potential problem. We have applied a synthetic biology approach to develop reporters that are capable of measuring transcription factor activity in vivo in real time. These synthetic reporters are comprised of a constitutive promoter with an operator site for the specific transcription factor immediately downstream. Thus, increasing transcription factor activity is measured as repression of expression of the transcription factor reporter. Measuring repression instead of activation avoids the complications of non-linear interactions between the transcription factor and RNA polymerase which differs at each promoter.
Results: Using these reporters, we show that a simple model is capable of determining the rules of integration for multiple transcriptional inputs at the four promoters of the arabinose catabolic pathway. Furthermore, we show that despite the complex and non-linear changes in cAMP-CRP activity in vivo during diauxic shift, the synthetic transcription factor reporters are capable of measuring real-time changes in transcription factor activity, and the simple model is capable of predicting the dynamic behaviour of the catabolic promoters.
Conclusions: Using a synthetic biology approach we show that the in vivo activity of transcription factors can be quantified without the need for measuring intracellular concentrations, binding affinities and reaction rates. Using measured transcription factor activity we show how different promoters can integrate common transcriptional inputs, resulting in distinct expression patterns. The data collected show that cAMP levels in vivo are dynamic and agree with observations showing that cAMP levels show a transient pulse during diauxic shift.
Figures
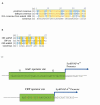
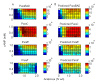
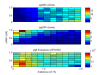
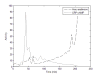
Similar articles
-
Design of CytR regulated, cAMP-CRP dependent class II promoters in Escherichia coli: RNA polymerase-promoter interactions modulate the efficiency of CytR repression.J Mol Biol. 1997 Mar 14;266(5):866-76. doi: 10.1006/jmbi.1996.0852. J Mol Biol. 1997. PMID: 9086266
-
Cautionary Notes on the Use of Arabinose- and Rhamnose-Inducible Expression Vectors in Pseudomonas aeruginosa.J Bacteriol. 2021 Jul 22;203(16):e0022421. doi: 10.1128/JB.00224-21. Epub 2021 Jul 22. J Bacteriol. 2021. PMID: 34096777 Free PMC article.
-
Vibrio vulnificus rpoS expression is repressed by direct binding of cAMP-cAMP receptor protein complex to its two promoter regions.J Biol Chem. 2008 Nov 7;283(45):30438-50. doi: 10.1074/jbc.M802219200. Epub 2008 Aug 18. J Biol Chem. 2008. PMID: 18713737 Free PMC article.
-
Dissecting the functional program of Escherichia coli promoters: the combined mode of action of Lac repressor and AraC activator.Nucleic Acids Res. 2001 Sep 15;29(18):3873-81. doi: 10.1093/nar/29.18.3873. Nucleic Acids Res. 2001. PMID: 11557820 Free PMC article.
-
Transcription activation by catabolite activator protein (CAP).J Mol Biol. 1999 Oct 22;293(2):199-213. doi: 10.1006/jmbi.1999.3161. J Mol Biol. 1999. PMID: 10550204 Review.
Cited by
-
Circuit-level input integration in bacterial gene regulation.Proc Natl Acad Sci U S A. 2013 Apr 23;110(17):7091-6. doi: 10.1073/pnas.1216091110. Epub 2013 Apr 9. Proc Natl Acad Sci U S A. 2013. PMID: 23572583 Free PMC article.
-
DNA is an antimicrobial component of neutrophil extracellular traps.PLoS Pathog. 2015 Jan 15;11(1):e1004593. doi: 10.1371/journal.ppat.1004593. eCollection 2015 Jan. PLoS Pathog. 2015. PMID: 25590621 Free PMC article.
-
Dissecting specific and global transcriptional regulation of bacterial gene expression.Mol Syst Biol. 2013 Apr 16;9:658. doi: 10.1038/msb.2013.14. Mol Syst Biol. 2013. PMID: 23591774 Free PMC article.
References
-
- Muller-Hill B. The lac operon: A Short History of a Genetic Paradigm. Berlin: Walter de Gruyter; 1996.
-
- Monod J. Recherches sur la croissance des cellules bacteriennes. Institute Pasteur, Acutalites scienctific et industrielles; 1942.
-
- Roseman S, Meadow ND. Signal transduction by the bacterial phosphotransferase system. Journal of Biological Chemistry. 1990;265:2993–2996. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

