The hidden duplication past of the plant pathogen Phytophthora and its consequences for infection
- PMID: 20525264
- PMCID: PMC2996974
- DOI: 10.1186/1471-2164-11-353
The hidden duplication past of the plant pathogen Phytophthora and its consequences for infection
Abstract
Background: Oomycetes of the genus Phytophthora are pathogens that infect a wide range of plant species. For dicot hosts such as tomato, potato and soybean, Phytophthora is even the most important pathogen. Previous analyses of Phytophthora genomes uncovered many genes, large gene families and large genome sizes that can partially be explained by significant repeat expansion patterns.
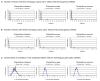
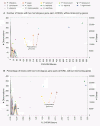
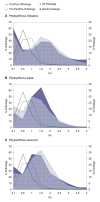
Results: Analysis of the complete genomes of three different Phytophthora species, using a newly developed approach, unveiled a large number of small duplicated blocks, mainly consisting of two or three consecutive genes. Further analysis of these duplicated genes and comparison with the known gene and genome duplication history of ten other eukaryotes including parasites, algae, plants, fungi, vertebrates and invertebrates, suggests that the ancestor of P. infestans, P. sojae and P. ramorum most likely underwent a whole genome duplication (WGD). Genes that have survived in duplicate are mainly genes that are known to be preferentially retained following WGDs, but also genes important for pathogenicity and infection of the different hosts seem to have been retained in excess. As a result, the WGD might have contributed to the evolutionary and pathogenic success of Phytophthora.
Conclusions: The fact that we find many small blocks of duplicated genes indicates that the genomes of Phytophthora species have been heavily rearranged following the WGD. Most likely, the high repeat content in these genomes have played an important role in this rearrangement process. As a consequence, the paucity of retained larger duplicated blocks has greatly complicated previous attempts to detect remnants of a large-scale duplication event in Phytophthora. However, as we show here, our newly developed strategy to identify very small duplicated blocks might be a useful approach to uncover ancient polyploidy events, in particular for heavily rearranged genomes.
Figures


References
-
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science (New York, NY) 2000;290(5493):972–977. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

