Network properties of human disease genes with pleiotropic effects
- PMID: 20525321
- PMCID: PMC2892460
- DOI: 10.1186/1752-0509-4-78
Network properties of human disease genes with pleiotropic effects
Abstract
Background: The ability of a gene to cause a disease is known to be associated with the topological position of its protein product in the molecular interaction network. Pleiotropy, in human genetic diseases, refers to the ability of different mutations within the same gene to cause different pathological effects. Here, we hypothesized that the ability of human disease genes to cause pleiotropic effects would be associated with their network properties.
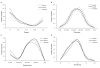
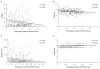
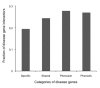
Results: Shared genes, with pleiotropic effects, were more central than specific genes that were associated with one disease, in the protein interaction network. Furthermore, shared genes associated with phenotypically divergent diseases (phenodiv genes) were more central than those associated with phenotypically similar diseases. Shared genes had a higher number of disease gene interactors compared to specific genes, implying higher likelihood of finding a novel disease gene in their network neighborhood. Shared genes had a relatively restricted tissue co-expression with interactors, contrary to specific genes. This could be a function of shared genes leading to pleiotropy. Essential and phenodiv genes had comparable connectivities and hence we investigated for differences in network attributes conferring lethality and pleiotropy, respectively. Essential and phenodiv genes were found to be intra-modular and inter-modular hubs with the former being highly co-expressed with their interactors contrary to the latter. Essential genes were predominantly nuclear proteins with transcriptional regulation activities while phenodiv genes were cytoplasmic proteins involved in signal transduction.
Conclusion: The properties of a disease gene in molecular interaction network determine its role in manifesting different and divergent diseases.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

