Intrinsic response of thoracic propriospinal neurons to axotomy
- PMID: 20525361
- PMCID: PMC2894843
- DOI: 10.1186/1471-2202-11-69
Intrinsic response of thoracic propriospinal neurons to axotomy
Abstract
Background: Central nervous system axons lack a robust regenerative response following spinal cord injury (SCI) and regeneration is usually abortive. Supraspinal pathways, which are the most commonly studied for their regenerative potential, demonstrate a limited regenerative ability. On the other hand, propriospinal (PS) neurons, with axons intrinsic to the spinal cord, have shown a greater regenerative response than their supraspinal counterparts, but remain relatively understudied in regards to spinal cord injury.
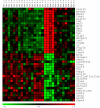
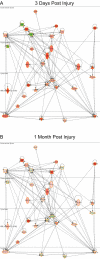
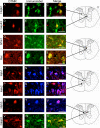
Results: Utilizing laser microdissection, gene-microarray, qRT-PCR, and immunohistochemistry, we focused on the intrinsic post-axotomy response of specifically labelled thoracic propriospinal neurons at periods from 3-days to 1-month following T9 spinal cord injury. We found a strong and early (3-days post injury, p.i) upregulation in the expression of genes involved in the immune/inflammatory response that returned towards normal by 1-week p.i. In addition, several regeneration associated and cell survival/neuroprotective genes were significantly up-regulated at the earliest p.i. period studied. Significant upregulation of several growth factor receptor genes (GFRa1, Ret, Lifr) also occurred only during the initial period examined. The expression of a number of pro-apoptotic genes up-regulated at 3-days p.i. suggest that changes in gene expression after this period may have resulted from analyzing surviving TPS neurons after the cell death of the remainder of the axotomized TPS neuronal population.
Conclusions: Taken collectively these data demonstrate that thoracic propriospinal (TPS) neurons mount a very dynamic response following low thoracic axotomy that includes a strong regenerative response, but also results in the cell death of many axotomized TPS neurons in the first week after spinal cord injury. These data also suggest that the immune/inflammatory response may have an important role in mediating the early strong regenerative response, as well as the apoptotic response, since expression of all of three classes of gene are up-regulated only during the initial period examined, 3-days post-SCI. The up-regulation in the expression of genes for several growth factor receptors during the first week post-SCI also suggest that administration of these factors may protect TPS neurons from cell death and maintain a regenerative response, but only if given during the early period after injury.
Figures



Similar articles
-
Long descending cervical propriospinal neurons differ from thoracic propriospinal neurons in response to low thoracic spinal injury.BMC Neurosci. 2010 Nov 23;11:148. doi: 10.1186/1471-2202-11-148. BMC Neurosci. 2010. PMID: 21092315 Free PMC article.
-
Effect of lesion proximity on the regenerative response of long descending propriospinal neurons after spinal transection injury.BMC Neurosci. 2019 Mar 18;20(1):10. doi: 10.1186/s12868-019-0491-y. BMC Neurosci. 2019. PMID: 30885135 Free PMC article.
-
Cell survival or cell death: differential vulnerability of long descending and thoracic propriospinal neurons to low thoracic axotomy in the adult rat.Neuroscience. 2011 Oct 27;194:359-71. doi: 10.1016/j.neuroscience.2011.05.052. Epub 2011 May 31. Neuroscience. 2011. PMID: 21645590
-
Transplants and neurotrophic factors increase regeneration and recovery of function after spinal cord injury.Prog Brain Res. 2002;137:257-73. doi: 10.1016/s0079-6123(02)37020-1. Prog Brain Res. 2002. PMID: 12440372 Review.
-
The Dorsal Column Lesion Model of Spinal Cord Injury and Its Use in Deciphering the Neuron-Intrinsic Injury Response.Dev Neurobiol. 2018 Oct;78(10):926-951. doi: 10.1002/dneu.22601. Epub 2018 May 11. Dev Neurobiol. 2018. PMID: 29717546 Free PMC article. Review.
Cited by
-
T12-L3 Nerve Transfer-Induced Locomotor Recovery in Rats with Thoracolumbar Contusion: Essential Roles of Sensory Input Rerouting and Central Neuroplasticity.Cells. 2023 Dec 8;12(24):2804. doi: 10.3390/cells12242804. Cells. 2023. PMID: 38132124 Free PMC article.
-
Long descending cervical propriospinal neurons differ from thoracic propriospinal neurons in response to low thoracic spinal injury.BMC Neurosci. 2010 Nov 23;11:148. doi: 10.1186/1471-2202-11-148. BMC Neurosci. 2010. PMID: 21092315 Free PMC article.
-
And yet it moves: Recovery of volitional control after spinal cord injury.Prog Neurobiol. 2018 Jan;160:64-81. doi: 10.1016/j.pneurobio.2017.10.004. Epub 2017 Nov 2. Prog Neurobiol. 2018. PMID: 29102670 Free PMC article. Review.
-
Required growth facilitators propel axon regeneration across complete spinal cord injury.Nature. 2018 Sep;561(7723):396-400. doi: 10.1038/s41586-018-0467-6. Epub 2018 Aug 29. Nature. 2018. PMID: 30158698 Free PMC article.
-
Identification of regenerative processes in neonatal spinal cord injury in the opossum (Monodelphis domestica): A transcriptomic study.J Comp Neurol. 2021 Apr 1;529(5):969-986. doi: 10.1002/cne.24994. Epub 2020 Aug 4. J Comp Neurol. 2021. PMID: 32710567 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

