Proteomic analysis of the fish pathogen Flavobacterium columnare
- PMID: 20525376
- PMCID: PMC2890538
- DOI: 10.1186/1477-5956-8-26
Proteomic analysis of the fish pathogen Flavobacterium columnare
Abstract
Background: Flavobacterium columnare causes columnaris disease in cultured and wild fish populations worldwide. Columnaris is the second most prevalent bacterial disease of commercial channel catfish industry in the United States. Despite its economic importance, little is known about the expressed proteins and virulence mechanisms of F. columnare. Here, we report the first high throughput proteomic analysis of F. columnare using 2-D LC ESI MS/MS and 2-DE MALDI TOF/TOF MS.
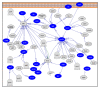
Results: Proteins identified in this study and predicted from the draft F. columnare genome were clustered into functional groups using clusters of orthologous groups (COGs), and their subcellular locations were predicted. Possible functional relations among the identified proteins were determined using pathway analysis. The total number of unique F. columnare proteins identified using both 2-D LC and 2-DE approaches was 621, of which 10.95% (68) were identified by both methods, while 77.29% (480) and 11.76% (73) were unique in 2-D LC and 2-DE, respectively. COG groupings and subcellular localizations were similar between our data set and proteins predicted from the whole genome. Twenty eight pathways were significantly represented in our dataset (P < 0.05).
Conclusion: Results from this study provide experimental evidence for many proteins that were predicted from the F. columnare genome annotation, and they should accelerate functional and comparative studies aimed at understanding virulence mechanisms of this important pathogen.
Figures



Similar articles
-
Type IX Secretion System Effectors and Virulence of the Model Flavobacterium columnare Strain MS-FC-4.Appl Environ Microbiol. 2022 Feb 8;88(3):e0170521. doi: 10.1128/AEM.01705-21. Epub 2021 Nov 24. Appl Environ Microbiol. 2022. PMID: 34818105 Free PMC article.
-
Proteome analysis of Edwardsiella ictaluri.Proteomics. 2009 Mar;9(5):1353-63. doi: 10.1002/pmic.200800652. Proteomics. 2009. PMID: 19253294
-
Putative roles for a rhamnose binding lectin in Flavobacterium columnare pathogenesis in channel catfish Ictalurus punctatus.Fish Shellfish Immunol. 2012 Oct;33(4):1008-15. doi: 10.1016/j.fsi.2012.08.018. Epub 2012 Aug 31. Fish Shellfish Immunol. 2012. PMID: 22960031
-
The Type IX Secretion System Is Required for Virulence of the Fish Pathogen Flavobacterium columnare.Appl Environ Microbiol. 2017 Nov 16;83(23):e01769-17. doi: 10.1128/AEM.01769-17. Print 2017 Dec 1. Appl Environ Microbiol. 2017. PMID: 28939608 Free PMC article.
-
Flavobacterium columnare infections in fish: the agent and its adhesion to the gill tissue.Verh K Acad Geneeskd Belg. 2002;64(6):421-30. Verh K Acad Geneeskd Belg. 2002. PMID: 12649933 Review.
Cited by
-
Mediation of Mucosal Immunoglobulins in Buccal Cavity of Teleost in Antibacterial Immunity.Front Immunol. 2020 Sep 23;11:562795. doi: 10.3389/fimmu.2020.562795. eCollection 2020. Front Immunol. 2020. PMID: 33072100 Free PMC article.
-
Phage-driven loss of virulence in a fish pathogenic bacterium.PLoS One. 2012 Dec 20;7(12):e53157. doi: 10.1371/journal.pone.0053157. Epub 2012 Dec 31. PLoS One. 2012. PMID: 23308090 Free PMC article.
-
Secreted Extracellular Products of Flavobacterium covae as Potential Immunogenic Factors for Protection against Columnaris Disease in Channel Catfish (Ictalurus punctatus).Pathogens. 2023 Jun 7;12(6):808. doi: 10.3390/pathogens12060808. Pathogens. 2023. PMID: 37375498 Free PMC article.
-
The role of denitrification genes in anaerobic growth and virulence of Flavobacterium columnare.J Appl Microbiol. 2021 Apr;130(4):1062-1074. doi: 10.1111/jam.14855. Epub 2020 Sep 30. J Appl Microbiol. 2021. PMID: 32955778 Free PMC article.
-
Speciation in caves: experimental evidence that permanent darkness promotes reproductive isolation.Biol Lett. 2011 Dec 23;7(6):909-12. doi: 10.1098/rsbl.2011.0237. Epub 2011 May 11. Biol Lett. 2011. PMID: 21561964 Free PMC article.
References
-
- Bernardet J, Segers P, Cancanneyt M, Berthe M, Kersters K, Vandamme P. Cutting a Gordian knot: emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriaceae, and proposal of Flavobacterium hydatis nom. nov. (Basonym, Cytophaga aquatilis Strohl and Tait 1978) Int J Syst Bacteriol. 1996;46:128–148. doi: 10.1099/00207713-46-1-128. - DOI
-
- Bernardet JF. Immunization with bacterial antigens: Flavobacterium and Flexibacter infections. Dev Biol Stand. 1997;90:179–188. - PubMed
-
- Plumb J. Health maintenance and microbial diseases of cultured fishes. Iowa State University Press, Ames, IA. 1999.
-
- Wakabayashi H. Effect of environmental conditions on the infectivity of Flexibacter columnaris to fish. J Fish Dis. 1991;14:279–290. doi: 10.1111/j.1365-2761.1991.tb00825.x. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

