Mechanical stretch up-regulates microRNA-26a and induces human airway smooth muscle hypertrophy by suppressing glycogen synthase kinase-3β
- PMID: 20525681
- PMCID: PMC2937966
- DOI: 10.1074/jbc.M110.101147
Mechanical stretch up-regulates microRNA-26a and induces human airway smooth muscle hypertrophy by suppressing glycogen synthase kinase-3β
Abstract
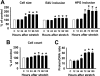
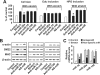
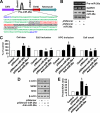
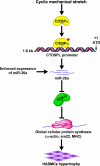
Airway smooth muscle hypertrophy is one of the hallmarks of airway remodeling in severe asthma. Several human diseases have been now associated with dysregulated microRNA (miRNA) expression. miRNAs are a class of small non-coding RNAs, which negatively regulate gene expression at the post-transcriptional level. Here, we identify miR-26a as a hypertrophic miRNA of human airway smooth muscle cells (HASMCs). We show that stretch selectively induces the transcription of miR-26a located in the locus 3p21.3 of human chromosome 3. The transcription factor CCAAT enhancer-binding protein α (C/EBPα) directly activates miR-26a expression through the transcriptional machinery upon stretch. Furthermore, stretch or enforced expression of miR-26a induces HASMC hypertrophy, and miR-26 knockdown reverses this effect, suggesting that miR-26a is a hypertrophic gene. We identify glycogen synthase kinase-3β (GSK-3β), an anti-hypertrophic protein, as a target gene of miR-26a. Luciferase reporter assays demonstrate that miR-26a directly interact with the 3'-untranslated repeat of the GSK-3β mRNA. Stretch or enforced expression of miR-26a attenuates the endogenous GSK-3β protein levels followed by the induction of HASMC hypertrophy. miR-26 knockdown reverses this effect, suggesting that miR-26a-induced hypertrophy occurs via its target gene GSK-3β. Overall, as a first time, our study unveils that miR-26a is a mechanosensitive gene, and it plays an important role in the regulation of HASMC hypertrophy.
Figures
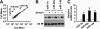





Similar articles
-
Desmin regulates airway smooth muscle hypertrophy through early growth-responsive protein-1 and microRNA-26a.J Biol Chem. 2011 Dec 16;286(50):43394-404. doi: 10.1074/jbc.M111.235127. Epub 2011 Sep 7. J Biol Chem. 2011. PMID: 21903578 Free PMC article.
-
Inhibition of glycogen synthase kinase-3beta is sufficient for airway smooth muscle hypertrophy.J Biol Chem. 2008 Apr 11;283(15):10198-207. doi: 10.1074/jbc.M800624200. Epub 2008 Feb 5. J Biol Chem. 2008. PMID: 18252708 Free PMC article.
-
Airway smooth muscle hyperplasia and hypertrophy correlate with glycogen synthase kinase-3(beta) phosphorylation in a mouse model of asthma.Am J Physiol Lung Cell Mol Physiol. 2009 Feb;296(2):L176-84. doi: 10.1152/ajplung.90376.2008. Epub 2008 Nov 14. Am J Physiol Lung Cell Mol Physiol. 2009. PMID: 19011050 Free PMC article.
-
MicroRNA-26a induces osteosarcoma cell growth and metastasis via the Wnt/β-catenin pathway.Oncol Lett. 2016 Feb;11(2):1592-1596. doi: 10.3892/ol.2015.4073. Epub 2015 Dec 31. Oncol Lett. 2016. PMID: 26893786 Free PMC article.
-
MicroRNA-26a in respiratory diseases: mechanisms and therapeutic potential.Mol Biol Rep. 2024 May 8;51(1):627. doi: 10.1007/s11033-024-09576-5. Mol Biol Rep. 2024. PMID: 38717532 Review.
Cited by
-
Mechanisms and therapeutic potential of microRNAs in hypertension.Drug Discov Today. 2015 Oct;20(10):1188-204. doi: 10.1016/j.drudis.2015.05.007. Epub 2015 May 21. Drug Discov Today. 2015. PMID: 26004493 Free PMC article. Review.
-
Decoding the non-coding RNAs in Alzheimer's disease.Cell Mol Life Sci. 2012 Nov;69(21):3543-59. doi: 10.1007/s00018-012-1125-z. Epub 2012 Sep 6. Cell Mol Life Sci. 2012. PMID: 22955374 Free PMC article. Review.
-
MiR-26a functions oppositely in osteogenic differentiation of BMSCs and ADSCs depending on distinct activation and roles of Wnt and BMP signaling pathway.Cell Death Dis. 2015 Aug 6;6(8):e1851. doi: 10.1038/cddis.2015.221. Cell Death Dis. 2015. PMID: 26247736 Free PMC article.
-
Aberrant DNA Methylation of Phosphodiesterase [corrected] 4D Alters Airway Smooth Muscle Cell Phenotypes.Am J Respir Cell Mol Biol. 2016 Feb;54(2):241-9. doi: 10.1165/rcmb.2015-0079OC. Am J Respir Cell Mol Biol. 2016. PMID: 26181301 Free PMC article.
-
The pivotal role of airway smooth muscle in asthma pathophysiology.J Allergy (Cairo). 2011;2011:742710. doi: 10.1155/2011/742710. Epub 2011 Dec 11. J Allergy (Cairo). 2011. PMID: 22220184 Free PMC article.
References
-
- Bartel D. P. (2004) Cell 116, 281–297 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

