Intercohort gene expression co-analysis reveals chemokine receptors as prognostic indicators in Ewing's sarcoma
- PMID: 20525755
- PMCID: PMC2905506
- DOI: 10.1158/1078-0432.CCR-10-0558
Intercohort gene expression co-analysis reveals chemokine receptors as prognostic indicators in Ewing's sarcoma
Abstract
Purpose: We report a novel analytic method, named intercohort co-analysis or Ican, which aids in the discovery of genes with predictive value for the progression or outcome of diseases from small-size cohorts. We tested this premise in Ewing's sarcoma (ES), a highly metastatic cancer of bone and soft tissues that lacks validated molecular metastasis and prognostic indicators.
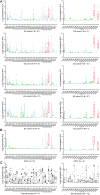
Experimental design: To uncover genes significantly expressed in ES patient subsets, we first determined a nonarbitrary gene expression significance cutoff based on expression levels in validated expressing and nonexpressing tissues. We next searched for genes that were consistently significantly expressed in several ES cohort and cell line datasets. Significantly expressed genes were independently validated by quantitative reverse transcription-PCR in an additional ES cohort.
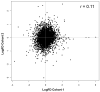
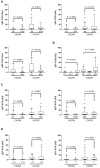
Results: Analysis of ES cohorts revealed marked intercohort gene expression variability. After filtering out the intercohort variability, CXCR4 and CXCR7 were found to be consistently associated with specific ES subsets. Pairwise analyses showed CXCR4 to correlate with ES metastases, and CXCR4 and CXCR7 to patient survival, but not with several other clinicopathological variables.
Conclusion: Ican is a powerful novel method to identifying genes consistently associated with particular disease states in cancers for which large cohorts are not available, currently the case of most cancers. We report for the first time that high CXCR4 expression preferentially associates with metastatic ES, and that of CXCR7 with poor patient survival.
Copyright 2010 AACR.
Figures
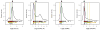

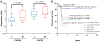
References
-
- Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nature medicine. 2002;8:816–24. - PubMed
-
- Varambally S, Yu J, Laxman B, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer cell. 2005;8:393–406. - PubMed
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. - PubMed
-
- Brown RC, Dwyer T, Kasten C, et al. Cohort profile: the International Childhood Cancer Cohort Consortium (I4C) Int J Epidemiol. 2007;36:724–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

