Using next-generation sequencing for high resolution multiplex analysis of copy number variation from nanogram quantities of DNA from formalin-fixed paraffin-embedded specimens
- PMID: 20525786
- PMCID: PMC2919738
- DOI: 10.1093/nar/gkq510
Using next-generation sequencing for high resolution multiplex analysis of copy number variation from nanogram quantities of DNA from formalin-fixed paraffin-embedded specimens
Abstract
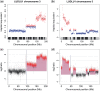
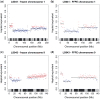
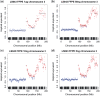
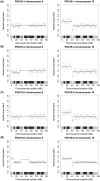
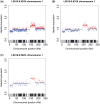
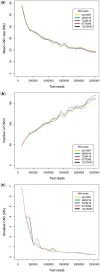
The use of next-generation sequencing technologies to produce genomic copy number data has recently been described. Most approaches, however, reply on optimal starting DNA, and are therefore unsuitable for the analysis of formalin-fixed paraffin-embedded (FFPE) samples, which largely precludes the analysis of many tumour series. We have sought to challenge the limits of this technique with regards to quality and quantity of starting material and the depth of sequencing required. We confirm that the technique can be used to interrogate DNA from cell lines, fresh frozen material and FFPE samples to assess copy number variation. We show that as little as 5 ng of DNA is needed to generate a copy number karyogram, and follow this up with data from a series of FFPE biopsies and surgical samples. We have used various levels of sample multiplexing to demonstrate the adjustable resolution of the methodology, depending on the number of samples and available resources. We also demonstrate reproducibility by use of replicate samples and comparison with microarray-based comparative genomic hybridization (aCGH) and digital PCR. This technique can be valuable in both the analysis of routine diagnostic samples and in examining large repositories of fixed archival material.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

