CcpA mediates proline auxotrophy and is required for Staphylococcus aureus pathogenesis
- PMID: 20525824
- PMCID: PMC2916365
- DOI: 10.1128/JB.00237-10
CcpA mediates proline auxotrophy and is required for Staphylococcus aureus pathogenesis
Abstract
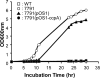
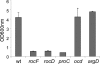
Human clinical isolates of Staphylococcus aureus, for example, strains Newman and N315, cannot grow in the absence of proline, albeit their sequenced genomes harbor genes for two redundant proline synthesis pathways. We show here that under selective pressure, S. aureus Newman generates proline-prototrophic variants at a frequency of 3 x 10(-6), introducing frameshift and missense mutations in ccpA or IS1811 insertions in ptsH, two regulatory genes that carry out carbon catabolite repression (CCR) in staphylococci and other Gram-positive bacteria. S. aureus Newman variants with mutations in rocF (arginase), rocD (ornithine aminotransferase), and proC (Delta(1)-pyrroline 5-carboxylate [P5C] reductase) are unable to generate proline-prototrophic variants, whereas a variant with a mutation in ocd (ornithine cyclodeaminase) is unaffected. Transposon insertion in ccpA also restored proline prototrophy. CcpA was shown to repress transcription of rocF and rocD, encoding the first two enzymes, but not of proC, encoding the third and final enzyme in the P5C reductase pathway. CcpA bound to the upstream regions of rocF and rocD but not to that of proC. CcpA's binding to the upstream regions was greatly enhanced by phosphorylated HPr. The CCR-mediated proline auxotrophy was lifted when nonpreferred carbohydrates were used as the sole carbon source. The ccpA mutant displayed reduced staphylococcal load and replication in a murine model of staphylococcal abscess formation, indicating that carbon catabolite repression presents an important pathogenesis strategy of S. aureus infections.
Figures






Similar articles
-
Staphylococcus aureus Does Not Synthesize Arginine from Proline under Physiological Conditions.J Bacteriol. 2022 Jun 21;204(6):e0001822. doi: 10.1128/jb.00018-22. Epub 2022 May 12. J Bacteriol. 2022. PMID: 35546540 Free PMC article.
-
CcpA regulates arginine biosynthesis in Staphylococcus aureus through repression of proline catabolism.PLoS Pathog. 2012;8(11):e1003033. doi: 10.1371/journal.ppat.1003033. Epub 2012 Nov 29. PLoS Pathog. 2012. PMID: 23209408 Free PMC article.
-
CcpA Affects Infectivity of Staphylococcus aureus in a Hyperglycemic Environment.Front Cell Infect Microbiol. 2017 May 9;7:172. doi: 10.3389/fcimb.2017.00172. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28536677 Free PMC article.
-
Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria?Mol Microbiol. 1995 Feb;15(3):395-401. doi: 10.1111/j.1365-2958.1995.tb02252.x. Mol Microbiol. 1995. PMID: 7540244 Review.
-
Carbon catabolite repression by the catabolite control protein CcpA in Staphylococcus xylosus.J Mol Microbiol Biotechnol. 2002 May;4(3):309-14. J Mol Microbiol Biotechnol. 2002. PMID: 11931563 Review.
Cited by
-
CcpA-independent glucose regulation of lactate dehydrogenase 1 in Staphylococcus aureus.PLoS One. 2013;8(1):e54293. doi: 10.1371/journal.pone.0054293. Epub 2013 Jan 14. PLoS One. 2013. PMID: 23342123 Free PMC article.
-
Mutation of L-2,3-diaminopropionic acid synthase genes blocks staphyloferrin B synthesis in Staphylococcus aureus.BMC Microbiol. 2011 Sep 9;11:199. doi: 10.1186/1471-2180-11-199. BMC Microbiol. 2011. PMID: 21906287 Free PMC article.
-
Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors.Proc Natl Acad Sci U S A. 2014 Sep 9;111(36):13187-92. doi: 10.1073/pnas.1407616111. Epub 2014 Aug 26. Proc Natl Acad Sci U S A. 2014. PMID: 25161283 Free PMC article.
-
mSphere of Influence: Metabolic redundancies enhance pathogenesis.mSphere. 2024 Jul 30;9(7):e0023924. doi: 10.1128/msphere.00239-24. Epub 2024 Jul 3. mSphere. 2024. PMID: 38958458 Free PMC article.
-
NanI Sialidase, CcpA, and CodY Work Together To Regulate Epsilon Toxin Production by Clostridium perfringens Type D Strain CN3718.J Bacteriol. 2015 Oct;197(20):3339-53. doi: 10.1128/JB.00349-15. Epub 2015 Aug 10. J Bacteriol. 2015. PMID: 26260460 Free PMC article.
References
-
- Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. - PubMed
-
- Bae, T., and O. Schneewind. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58-63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

