Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense
- PMID: 20525853
- PMCID: PMC2910984
- DOI: 10.1105/tpc.109.070961
Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense
Abstract
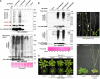
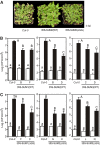
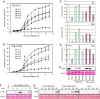
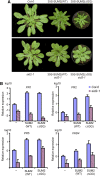
Posttranslational modifications allow dynamic and reversible changes to protein function. In Arabidopsis thaliana, a small gene family encodes paralogs of the small ubiquitin-like posttranslational modifier. We studied the function of these paralogs. Single mutants of the SUM1 and SUM2 paralogs do not exhibit a clear phenotype. However, the corresponding double knockdown mutant revealed that SUM1 and SUM2 are essential for plant development, floral transition, and suppression of salicylic acid (SA)-dependent defense responses. The SUM1 and SUM2 genes are constitutively expressed, but their spatial expression patterns do not overlap. Tight transcriptional regulation of these two SUM genes appears to be important, as overexpression of either wild-type or conjugation-deficient mutants resulted in activation of SA-dependent defense responses, as did the sum1 sum2 knockdown mutant. Interestingly, expression of the paralog SUM3 is strongly and widely induced by SA and by the defense elicitor Flg22, whereas its expression is otherwise low and restricted to a few specific cell types. Loss of SUM3 does not result in an aberrant developmental phenotype except for late flowering, while SUM3 overexpression causes early flowering and activates plant defense. Apparently, SUM3 promotes plant defense downstream of SA, while SUM1 and SUM2 together prevent SA accumulation in noninfected plants.
Figures




Similar articles
-
The Arabidopsis thaliana At4g13040 gene, a unique member of the AP2/EREBP family, is a positive regulator for salicylic acid accumulation and basal defense against bacterial pathogens.J Plant Physiol. 2014 Jun 15;171(10):860-7. doi: 10.1016/j.jplph.2013.12.015. Epub 2014 Mar 5. J Plant Physiol. 2014. PMID: 24612849
-
Arabidopsis GH3-LIKE DEFENSE GENE 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae.Plant J. 2007 Jul;51(2):234-46. doi: 10.1111/j.1365-313X.2007.03130.x. Epub 2007 May 23. Plant J. 2007. PMID: 17521413
-
Antagonism between SUMO1/2 and SUMO3 regulates SUMO conjugate levels and fine-tunes immunity.J Exp Bot. 2021 Sep 30;72(18):6640-6658. doi: 10.1093/jxb/erab296. J Exp Bot. 2021. PMID: 34145454
-
The expression patterns of AtBSMT1 and AtSAGT1 encoding a salicylic acid (SA) methyltransferase and a SA glucosyltransferase, respectively, in Arabidopsis plants with altered defense responses.Mol Cells. 2009 Aug 31;28(2):105-9. doi: 10.1007/s10059-009-0108-x. Epub 2009 Jul 30. Mol Cells. 2009. PMID: 19669626
-
Dissection of salicylic acid-mediated defense signaling networks.Plant Signal Behav. 2009 Aug;4(8):713-7. doi: 10.4161/psb.4.8.9173. Epub 2009 Aug 3. Plant Signal Behav. 2009. PMID: 19820324 Free PMC article. Review.
Cited by
-
Highly sensitive site-specific SUMOylation proteomics in Arabidopsis.Nat Plants. 2024 Sep;10(9):1330-1342. doi: 10.1038/s41477-024-01783-z. Epub 2024 Sep 18. Nat Plants. 2024. PMID: 39294263
-
Insights into the transcriptional and post-transcriptional regulation of the rice SUMOylation machinery and into the role of two rice SUMO proteases.BMC Plant Biol. 2018 Dec 12;18(1):349. doi: 10.1186/s12870-018-1547-3. BMC Plant Biol. 2018. PMID: 30541427 Free PMC article.
-
Analysis of Small Ubiquitin-Like Modifier (SUMO) Targets Reflects the Essential Nature of Protein SUMOylation and Provides Insight to Elucidate the Role of SUMO in Plant Development.Plant Physiol. 2015 Oct;169(2):1006-17. doi: 10.1104/pp.15.01014. Epub 2015 Aug 28. Plant Physiol. 2015. PMID: 26320229 Free PMC article.
-
SCE1, the SUMO-conjugating enzyme in plants that interacts with NIb, the RNA-dependent RNA polymerase of Turnip mosaic virus, is required for viral infection.J Virol. 2013 Apr;87(8):4704-15. doi: 10.1128/JVI.02828-12. Epub 2013 Jan 30. J Virol. 2013. PMID: 23365455 Free PMC article.
-
Sugarcane genes differentially expressed in response to Puccinia melanocephala infection: identification and transcript profiling.Plant Cell Rep. 2012 May;31(5):955-69. doi: 10.1007/s00299-011-1216-6. Epub 2012 Jan 3. Plant Cell Rep. 2012. PMID: 22212461
References
-
- Anckar J., Sistonen L. (2007). SUMO: Getting it on. Biochem. Soc. Trans. 35: 1409–1413 - PubMed
-
- Baurle I., Dean C. (2006). The timing of developmental transitions in plants. Cell 125: 655–664 - PubMed
-
- Bernier-Villamor V., Sampson D.A., Matunis M.J., Lima C.D. (2002). Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108: 345–356 - PubMed
-
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

