Host association of Campylobacter genotypes transcends geographic variation
- PMID: 20525862
- PMCID: PMC2916502
- DOI: 10.1128/AEM.00124-10
Host association of Campylobacter genotypes transcends geographic variation
Abstract
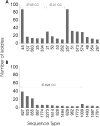
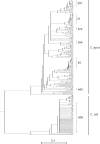
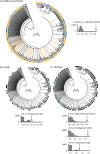
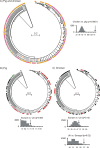
Genetic attribution of bacterial genotypes has become a major tool in the investigation of the epidemiology of campylobacteriosis and has implicated retail chicken meat as the major source of human infection in several countries. To investigate the robustness of this approach to the provenance of the reference data sets used, a collection of 742 Campylobacter jejuni and 261 Campylobacter coli isolates obtained from United Kingdom-sourced chicken meat was established and typed by multilocus sequence typing. Comparative analyses of the data with those from other isolates sourced from a variety of host animals and countries were undertaken by genetic attribution, genealogical, and population genetic approaches. The genotypes from the United Kingdom data set were highly diverse, yet structured into sequence types, clonal complexes, and genealogical groups very similar to those seen in chicken isolates from the Netherlands, the United States, and Senegal, but more distinct from isolates obtained from ruminant, swine, and wild bird sources. Assignment analyses consistently grouped isolates from different host animal sources regardless of geographical source; these associations were more robust than geographic associations across isolates from three continents. We conclude that, notwithstanding the high diversity of these pathogens, there is a strong signal of association of multilocus genotypes with particular hosts, which is greater than the geographic signal. These findings are consistent with local and international transmission of host-associated lineages among food animal species and provide a foundation for further improvements in genetic attribution.
Figures


References
-
- Achtman, M. 2008. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu. Rev. Microbiol. 62:53-70. - PubMed
-
- Best, E. L., N. B. Powell, C. Swift, K. A. Grant, and J. A. Frost. 2003. Applicability of a rapid duplex real-time PCR assay for speciation of Campylobacter jejuni and Campylobacter coli directly from culture plates. FEMS Microbiol. Lett. 229:237-241. - PubMed
-
- Bolton, F. J., D. R. A. Wareing, M. B. Skirrow, and D. N. Hutchinson. 1992. Identification and biotyping of campylobacters, p. 151-161. In R. G. Board, D. Jones, and F. A. Skinner (ed.), Identification methods in applied and environmental microbiology. Blackwell Scientific Publications, Ltd., London, United Kingdom.
-
- Buzby, J. C., and T. Roberts. 1997. Economic costs and trade impacts of microbial foodbourne illness. World Health Stat. Q. 50:57-66. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

