RTX proteins: a highly diverse family secreted by a common mechanism
- PMID: 20528947
- PMCID: PMC3034196
- DOI: 10.1111/j.1574-6976.2010.00231.x
RTX proteins: a highly diverse family secreted by a common mechanism
Erratum in
-
Correction to: RTX proteins: A highly diverse family secreted by a common mechanism.FEMS Microbiol Rev. 2023 Jul 5;47(4):fuad024. doi: 10.1093/femsre/fuad024. FEMS Microbiol Rev. 2023. PMID: 37436822 Free PMC article. No abstract available.
Abstract
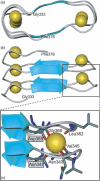
Repeats-in-toxin (RTX) exoproteins of Gram-negative bacteria form a steadily growing family of proteins with diverse biological functions. Their common feature is the unique mode of export across the bacterial envelope via the type I secretion system and the characteristic, typically nonapeptide, glycine- and aspartate-rich repeats binding Ca(2+) ions. In this review, we summarize the current state of knowledge on the organization of rtx loci and on the biological and biochemical activities of therein encoded proteins. Applying several types of bioinformatic screens on the steadily growing set of sequenced bacterial genomes, over 1000 RTX family members were detected, with the biological functions of most of them remaining to be characterized. Activities of the so far characterized RTX family members are then discussed and classified according to functional categories, ranging from the historically first characterized pore-forming RTX leukotoxins, through the large multifunctional enzymatic toxins, bacteriocins, nodulation proteins, surface layer proteins, up to secreted hydrolytic enzymes exhibiting metalloprotease or lipase activities of industrial interest.
Figures
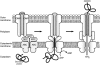


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

