Active site plasticity revealed from the structure of the enterobacterial N-ribohydrolase RihA bound to a competitive inhibitor
- PMID: 20529317
- PMCID: PMC2898832
- DOI: 10.1186/1472-6807-10-14
Active site plasticity revealed from the structure of the enterobacterial N-ribohydrolase RihA bound to a competitive inhibitor
Abstract
Background: Pyrimidine-preferring N-ribohydrolases (CU-NHs) are a class of Ca2+-dependent enzymes that catalyze the hydrolytic cleavage of the N-glycosidic bond in pyrimidine nucleosides. With the exception of few selected organisms, their physiological relevance in prokaryotes and eukaryotes is yet under investigation.
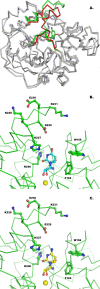
Results: Here, we report the first crystal structure of a CU-NH bound to a competitive inhibitor, the complex between the Escherichia coli enzyme RihA bound to 3, 4-diaminophenyl-iminoribitol (DAPIR) to a resolution of 2.1 A. The ligand can bind at the active site in two distinct orientations, and the stabilization of two flexible active site regions is pivotal to establish the interactions required for substrate discrimination and catalysis.
Conclusions: A comparison with the product-bound RihA structure allows a rationalization of the structural rearrangements required for an enzymatic catalytic cycle, highlighting a substrate-assisted cooperative motion, and suggesting a yet overlooked role of the conserved His82 residue in modulating product release. Differences in the structural features of the active sites in the two homologous CU-NHs RihA and RihB from E. coli provide a rationale for their fine differences in substrate specificity. These new findings hint at a possible role of CU-NHs in the breakdown of modified nucleosides derived from RNA molecules.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

