Localization of the two tropomyosin-binding sites of troponin T
- PMID: 20529660
- PMCID: PMC2904419
- DOI: 10.1016/j.abb.2010.06.001
Localization of the two tropomyosin-binding sites of troponin T
Abstract
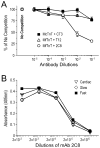
Troponin T (TnT) binds to tropomyosin (Tm) to anchor the troponin complex in the thin filament, and it thus serves as a vital link in the Ca(2+) regulation of striated muscle contraction. Pioneer work three decades ago determined that the T1 and T2 chymotryptic fragments of TnT each contains a Tm-binding site. A more precise localization of the two Tm-binding sites of TnT remains to be determined. In the present study, we tested serial deletion constructs of TnT and carried out monoclonal antibody competition experiments to show that the T1 region Tm-binding site involves mainly a 39 amino acids segment in the N-terminal portion of the conserved middle region of TnT. We further employed another set of TnT fragments to locate the T2 region Tm-binding site to a segment of 25 amino acids near the beginning of the T2 fragment. The localization of the two Tm-binding sites of TnT provided new information for the structure-function relationship of TnT and the anchoring of troponin complex on muscle thin filament.
2010 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
N-Terminal Hypervariable Region of Muscle Type Isoforms of Troponin T Differentially Modulates the Affinity of Tropomyosin-Binding Site 1.Biochemistry. 2015 Jun 23;54(24):3822-30. doi: 10.1021/acs.biochem.5b00348. Epub 2015 Jun 10. Biochemistry. 2015. PMID: 26024675
-
Cooperative interaction between developmentally regulated troponin T and tropomyosin isoforms in the absence of F-actin.J Biol Chem. 2000 Aug 25;275(34):26089-95. doi: 10.1074/jbc.M910360199. J Biol Chem. 2000. PMID: 10844003
-
Conformational modulation of slow skeletal muscle troponin T by an NH(2)-terminal metal-binding extension.Am J Physiol Cell Physiol. 2000 Oct;279(4):C1067-77. doi: 10.1152/ajpcell.2000.279.4.C1067. Am J Physiol Cell Physiol. 2000. PMID: 11003587
-
Evolution, Regulation, and Function of N-terminal Variable Region of Troponin T: Modulation of Muscle Contractility and Beyond.Int Rev Cell Mol Biol. 2016;321:1-28. doi: 10.1016/bs.ircmb.2015.09.002. Epub 2015 Nov 4. Int Rev Cell Mol Biol. 2016. PMID: 26811285 Review.
-
Structural interactions responsible for the assembly of the troponin complex on the muscle thin filament.Cell Struct Funct. 1997 Feb;22(1):219-23. doi: 10.1247/csf.22.219. Cell Struct Funct. 1997. PMID: 9113410 Review.
Cited by
-
FRET study of the structural and kinetic effects of PKC phosphomimetic cardiac troponin T mutants on thin filament regulation.Arch Biochem Biophys. 2014 May 15;550-551:1-11. doi: 10.1016/j.abb.2014.03.013. Epub 2014 Apr 5. Arch Biochem Biophys. 2014. PMID: 24708997 Free PMC article.
-
Novel autosomal dominant TNNT1 mutation causing nemaline myopathy.Mol Genet Genomic Med. 2017 Nov;5(6):678-691. doi: 10.1002/mgg3.325. Epub 2017 Aug 21. Mol Genet Genomic Med. 2017. PMID: 29178646 Free PMC article.
-
The highly conserved C-terminal end segment of troponin T binds tropomyosin and actin to function in modulating contractile kinetics.Proc Natl Acad Sci U S A. 2025 Jul 8;122(27):e2507107122. doi: 10.1073/pnas.2507107122. Epub 2025 Jul 1. Proc Natl Acad Sci U S A. 2025. PMID: 40591592
-
Proof of Principle that Molecular Modeling Followed by a Biophysical Experiment Can Develop Small Molecules that Restore Function to the Cardiac Thin Filament in the Presence of Cardiomyopathic Mutations.ACS Omega. 2019 Apr 30;4(4):6492-6501. doi: 10.1021/acsomega.8b03340. Epub 2019 Apr 9. ACS Omega. 2019. PMID: 31342001 Free PMC article.
-
Disrupted mechanobiology links the molecular and cellular phenotypes in familial dilated cardiomyopathy.Proc Natl Acad Sci U S A. 2019 Sep 3;116(36):17831-17840. doi: 10.1073/pnas.1910962116. Epub 2019 Aug 19. Proc Natl Acad Sci U S A. 2019. PMID: 31427533 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

