Robust super-resolution volume reconstruction from slice acquisitions: application to fetal brain MRI
- PMID: 20529730
- PMCID: PMC3694441
- DOI: 10.1109/TMI.2010.2051680
Robust super-resolution volume reconstruction from slice acquisitions: application to fetal brain MRI
Abstract
Fast magnetic resonance imaging slice acquisition techniques such as single shot fast spin echo are routinely used in the presence of uncontrollable motion. These techniques are widely used for fetal magnetic resonance imaging (MRI) and MRI of moving subjects and organs. Although high-quality slices are frequently acquired by these techniques, inter-slice motion leads to severe motion artifacts that are apparent in out-of-plane views. Slice sequential acquisitions do not enable 3-D volume representation. In this study, we have developed a novel technique based on a slice acquisition model, which enables the reconstruction of a volumetric image from multiple-scan slice acquisitions. The super-resolution volume reconstruction is formulated as an inverse problem of finding the underlying structure generating the acquired slices. We have developed a robust M-estimation solution which minimizes a robust error norm function between the model-generated slices and the acquired slices. The accuracy and robustness of this novel technique has been quantitatively assessed through simulations with digital brain phantom images as well as high-resolution newborn images. We also report here successful application of our new technique for the reconstruction of volumetric fetal brain MRI from clinically acquired data.
Figures






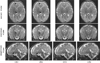



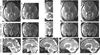
References
-
- Prayer D, Brugger P, Prayer L. Fetal MRI: techniques and protocols. Pediatric Radiology. 2004 Sep;vol. 34(no. 9):685–693. - PubMed
-
- Huppert BJ, Brandt KR, Ramin KD, King BF. Single-shot fast spin-echo MR imaging of the fetus: A pictorial essay. Radiographics. 1999;vol. 19(no. suppl 1):S215–S227. - PubMed
-
- Rutherford MA. Magnetic resonance imaging of the fetal brain. Current Opinion in Obstetrics and Gynecology. 2009;vol. 21(no. 2):180–186. - PubMed
-
- Vignaux OB, Augui J, Coste J, Argaud C, Le Roux P, Carlier PG, Duboc D, Legmann P. Comparison of single-shot fast spin-echo and conventional spin-echo sequences for MR imaging of the heart: initial experience. Radiology. 2001;vol. 219(no. 2):545–550. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

