Activity-based protein profiling identifies a host enzyme, carboxylesterase 1, which is differentially active during hepatitis C virus replication
- PMID: 20530478
- PMCID: PMC2919124
- DOI: 10.1074/jbc.M110.135483
Activity-based protein profiling identifies a host enzyme, carboxylesterase 1, which is differentially active during hepatitis C virus replication
Abstract
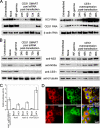
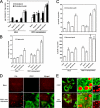
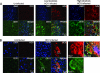
Hepatitis C virus (HCV) relies on many interactions with host cell proteins for propagation. Successful HCV infection also requires enzymatic activity of host cell enzymes for key post-translational modifications. To identify such enzymes, we have applied activity-based protein profiling to examine the activity of serine hydrolases during HCV replication. Profiling of hydrolases in Huh7 cells replicating HCV identified CES1 (carboxylesterase 1) as a differentially active enzyme. CES1 is an endogenous liver protein involved in processing of triglycerides and cholesterol. We observe that CES1 expression and activity were altered in the presence of HCV. The knockdown of CES1 with siRNA resulted in lower levels of HCV replication, and up-regulation of CES1 was observed to favor HCV propagation, implying an important role for this host cell protein. Experiments in HCV JFH1-infected cells suggest that CES1 facilitates HCV release because less intracellular HCV core protein was observed, whereas HCV titers remained high. CES1 activity was observed to increase the size and density of lipid droplets, which are necessary for the maturation of very low density lipoproteins, one of the likely vehicles for HCV release. In transgenic mice containing human-mouse chimeric livers, HCV infection also correlates with higher levels of endogenous CES1, providing further evidence that CES1 has an important role in HCV propagation.
Figures


References
-
- Lindenbach B. D., Rice C. M. (2005) Nature 436, 933–938 - PubMed
-
- Chisari F. V. (2005) Nature 436, 930–932 - PubMed
-
- Moradpour D., Penin F., Rice C. M. (2007) Nat. Rev. Microbiol. 5, 453–463 - PubMed
-
- Sagan S. M., Rouleau Y., Leggiadro C., Supekova L., Schultz P. G., Su A. I., Pezacki J. P. (2006) Biochem. Cell Biol. 84, 67–79 - PubMed
-
- Pezacki J. P., Singaravelu R., Lyn R. K. (2010) Mol. Biosys. 6, 1131–1142 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

